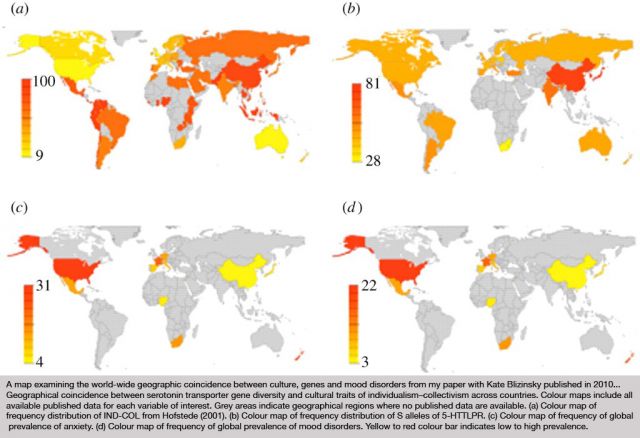
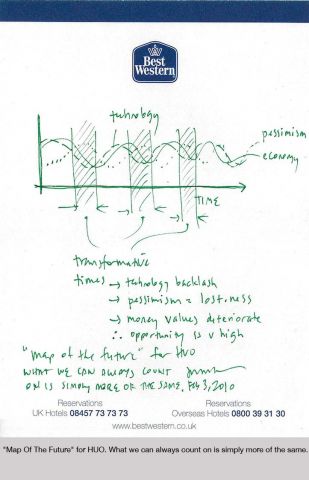
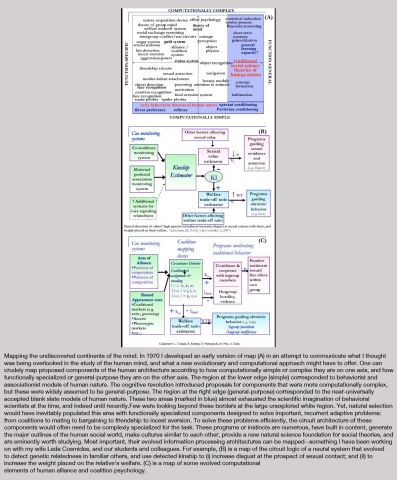
On Sunday, October 16th, Edge, at the invitation of London's leading curator, and long-time collaborator, Hans Ulrich Obrist (HUO), co-director of the Serpentine Gallery, participated in The Serpentine Gallery Garden Marathon, the sixth in the Gallery’s acclaimed Marathon series. The Garden Marathon explored the concept of the garden.
 As Obrist noted,
As Obrist noted,
"A product of the creative encounter between the man-made and the natural, between order and disorder, the garden can offer productive metaphors for the interactions between human life and time, care, thought or space."
"The event is directly inspired by the Serpentine Gallery Pavilion 2011, designed by Peter Zumthor. The encounter of architecture and garden creates a contemplative space that is both set within – and meditatively separated from – the wider surroundings of Kensington Gardens.
"Participations will range from the fields of horticulture, design and architecture to explore the creation of gardens and their spatial, urban and scientific importance, through to works by artists and readings by poets and writers exploring the significance of the garden in our experience of the world."
[Photo: Stefano Boeri, Vertical Forest, © Stefano Boeri]
Other recent collaborations between Edge and HUO have included "What is your Formula, Your Equation, Your Algorithm: Formulae for the 21st Century" in 2007. "Maps for the 21st Century" in 2010. HUO and I have also had the pleasure of writing about each other. See "Brockman's Taste for Science, or how to entertain the smartest people" by Hans Ulrich Obrist, and "A Rule of the Game: A Talk with Hans Ulrich Obrist on Edge.
INFORMATION GARDENS consisted of talks by Mark Pagel, Jennifer Jacquet, and Brian Eno.

John Brockman In Conversation with:
Mark Pagel, Cities as Gardens
"Up until 10,000 years ago there were no permanent settlements and all human groups lived by hunting and gathering. Then agriculture was discovered and everything changed. Now a small number of people could supply food for the rest and the first cities arose. Every since that time there has been a steady movement of people out of our original arcadia and into cities, such that now over half the world lives in them. But why given that cities have historically been targets of attack and places of crime and where diseases fester and spread? The answer is that cities have acted as gardens of our prosperity, creativity and innovations and their continued existence is vital to fitting the projected 9 billion people onto this planet. Surprisingly, they are the new 'green centres' of the world."
MARK PAGEL is a Fellow of the Royal Society and Professor of Evolutionary Biology; Head of the Evolution Laboratory at the University of Reading; Author Oxford Encyclopaedia of Evolution; co-author of The Comparative Method in Evolutionary Biology. Forthcoming book Wired for Culture: Origins of the Human Social Mind.

Jennifer Jacquet, Shame Totem v.2.0
"Throughout the 19th century, native tribes that spanned the north coast of North America erected shame totem poles to signal to the community that certain individuals or groups had transgressed. This art is resurrected with a modernized, garish, digitally rendered 3-D shame pole to represent the most shameful corporations – chosen with the assistance of 500 people based in the U.S. who surveyed about the corporations that have most negatively affected society. The talk will describe the relationship between gardens and shame, a historical view on shame totems, the specific concept for this work, and details of its creation."
JENNIFER JACQUET is a Postdoctoral Researcher, Fisheries Centre/Department of Mathematics, University of British Columbia; Research interests incude environmental sustainability (particularly fish), the evolution and function of guilt, honor, and shame, and the role of information technology in shaping environmental action.

Brian Eno, Composers as Gardeners
"My topic is the shift from 'architect' to 'gardener', where 'architect' stands for 'someone who carries a full picture of the work before it is made', to 'gardener' standing for 'someone who plants seeds and waits to see exactly what will come up'. I will argue that today's composer are more frequently 'gardeners' than 'architects' and, further, that the 'composer as architect' metaphor was a transitory historical blip."
Daniel Kahneman, Martin Nowak, Steven Pinker, Leda Cosmides, Michael Gazzaniga, Elaine Pagels
"We'd certainly be better off if everyone sampled the fabulous Edge symposium, which, like the best in science, is modest and daring all at once." — David Brooks, New York Times column
In July, Edge held its annual Master Class in Napa, California, on the theme: "The Science of Human Nature": Princeton psychologist Daniel Kahneman on the marvels and the flaws of intuitive thinking; Harvard mathematical biologist Martin Nowak on the evolution of cooperation; Harvard psychologist Steven Pinker on the history of violence; UC-Santa Barbara evolutionary psychologist Leda Cosmides on the architecture of motivation; UC-Santa Barbara neuroscientist Michael Gazzaniga on neuroscience and the law; and Princeton religious historian Elaine Pagels on The Book of Revelation. In the coming weeks we will publish the complete video, audio, and texts. For publication schedule and details, see below.
Spring Mountain Vineyard, St. Helena, Napa, CA
Friday July 15 to Sunday, July 17th

DANIEL KAHNEMAN: "THE MARVELS AND THE FLAWS OF INTUITIVE THINKING"
The power of settings, priming, and unconscious thinking, all are a major change in psychology. I can't think of a bigger change in my lifetime. You were asking what's exciting? That's exciting, to me.

Eugene Higgins Professor of Psychology, Princeton University; Recipient, the 2002 Nobel Prize in Economic Sciences; Author, Thinking Fast and Slow (forthcoming, October 25th). Daniel Kahneman's Edge Bio Page
[Continue to Daniel Kahneman's Edge Master Class]

MARTIN NOWAK: "THE EVOLUTION OF COOPERATION"
Why has cooperation, not competition, always been the key to the evolution of complexity?

Mathematical Biologist, Game Theorist; Professor of Biology and Mathematics, Director, Center for Evolutionary Dynamics, Harvard University; Author, SuperCooperators: Altruism, Evolution, and Why We Need Each Other to Succeed. Martin Nowak's Edge Bio Page
[Continue to Martin Nowak's Edge Master Class]

STEVEN PINKER: "A HISTORY OF VIOLENCE"
What may be the most important thing that has ever happened in human history is that violence has gone down, by dramatic degrees, and in many dimensions all over the world and in many spheres of behavior: genocide, war, human sacrifice, torture, slavery, and the treatment of racial minorities, women, children, and animals.
Harvard College Professor and Johnstone Family Professor of Psychology; Harvard University. Author, The Language Instinct, How the Mind Works, and The Better Angels Of Our Nature: How Violence Has Declined (forthcoming, October 4th). Steven Pinker's Edge Bio Page
[Continue to Steven Pinker's Edge Master Class]

LEDA COSMIDES: "THE ARCHITECTURE OF MOTIVATION"
Recent research concerning the welfare of others, etc. affects not only how to think about certain emotions, but also overturns how most models of reciprocity and exchange, with implications about how people think about modern markets, political systems, and societies. What are these new approaches to human motivation?

Professor of Psychology and Co-director (with John Tooby) of Center for Evolutionary Psychology at the University of California, Santa Barbara. Leda Cosmides's Edge Bio Page
[Continue to Leda Cosmides's Edge Master Class]

MICHAEL GAZZANIGA: "NEUROSCIENCE AND JUSTICE"
Asking the fundamental question of modern life. In an enlightened world of scientific understandings of first causes, we must ask: are we free, morally responsible agents or are we just along for the ride?

Neuroscientist; Professor of Psychology & Director, SAGE Center for the Study of Mind, University of California, Santa Barbara; Human: Who's In Charge? (forthcoming, November 15th). Michael Gazzaniga's Edge Bio Page
[Continue to Michael Gazzaniga's Edge Master Class]

ELAINE PAGELS: "THE BOOK OF REVELATION: PROPHECY AND POLITICS"
Why is religion still alive? Why are people still engaged in old folk takes and mythological stories — even those without rational and ethical foundations.

Harrington Spear Paine Professor of Religion, Princeton University; Author The Gnostic Gospels; Beyond Belief; and Revelations: Visions, Prophecy, and Politics in the Book of Revelation (forthcoming, March 6, 2012). Elaine Pagels's Edge Bio Page
[Continue to Elaine Pagels's Edge Master Class]
"Open-minded, free ranging, intellectually playful ... an unadorned pleasure in curiosity, a collective expression of wonder at the living and inanimate world ... an ongoing and thrilling colloquium." — Ian McEwan in The Telegraph

Villa Miravalle at Spring Mountain
The Edge Master Class 2011 was held at Villa Miravelle at Spring Mountain Vineyard in St. Helena, California.
Built 1884 in Saint Helena, CA, by Mexican-American Tiburcio Parrott, the majestic residence dominates the surrounding vineyards and includes spires, wraparound verandas, a conservatory, a grand stone tower, massive front double doors with exquisite stained glass, and a six-story high cupola. Miravalle was designed by architect Albert Schroepfer, who had designed acclaimed structures at Inglenook and Beringer Wineries, and San Fransisco's Orpheum Theatre. ... Tiburcio died within ten years, and Miravalle remained empty for the next seventy. In 1974 Spring Mountain Vineyard and winery were established on the surrounding property.
The Vineyard was bought by Edge member Jacqui (Jacob) Eli Safra in 1992, after which he consolidated several properties into the current 900-acre property, the largest contiguous vineyard in Napa. Safra, a Swiss investor, is a descendant of the Lebanon-Swiss Jewish Safra banking family. In addition to Spring Mountain Vineyards, his other investments include Encyclopædia Britannica and Merriam-Webster. The entire Edge community wishes to thank him for his thoughtfuness and generosity. And we wish to express our appreciation to General Manager George Peterson, and Customer Relationship Director Leah Smith for their help in organizing a memorable weekend.
George Dyson, Stewart Brand, John Brockman, Kevin Kelly
This year, Edge@DLD is presenting a session with three of the original members of Edge who year in and year out provide the core sounding board for the ideas and information we present to the public. I refer to them in private correspondance as "The Council"). Every year, beginning late summer, I consult with Stewart Brand, Kevin Kelly, and George Dyson, and together we create the Edge Annual Question which Edge has been asking for the past fourteen years.
Stewart Brand is the founder and editor of Whole Earth Catalog and author of Whole Earth Discipline; Kevin Kelly helped to launch Wiredin 1993 and is the author of Out of Control and What Technology Wants; and George Dyson, a science historian, is the author of Darwin Among the Machines and the forthcoming Turing's Cathedral.
This is fourth year that Edge has been invited to collaborate with DLD (Digitlal-Life-Day) in Munich. In 2008 genomics researcher the Edge Conversation featured J. Craig Venter and biologist Richard Dawkins on "Life: A Gene-Centric View". In 2009, psychologist and Nobel Laureate Daniel Kahneman and philosophical essayist & researcher Nassim Nicholas Taleb held forth on "Reflections On A Crisis." In 2010 the conversation included computer scientist David Gelernter, Feuilleton Editor (Sueddeutsche Zeitung) Andrian Kreye, and Feuilleton Editor & Co-Publisher (Frankfurter Allgemeine Zeitung) Frank Schirrmacher, who explored the idea of "The Informavore".
— JB
[1-3:24] Introduction: Andrian Kreye
[3:24-6:07] John Brockman, Moderator
[6:07-21:55] Stewart Brand
[21:55-21:40] Kevin Kelly
[21:40-51:05] George Dyson
Continue to transcripts and video of the discussion:
Edge@DLD
Stewart Brand, Kevin Kelly & George Dyson: An Edge Conversation In Munich

"The dinner party was a microcosm of a newly dominant sector of American business. "
— Gary Wolf, Wired
[Photos by Nathan Myhrvold]
We make a mistake when we think of cancer as a noun. It is not something you have, it is something you do. Your body is probably cancering all the time. What keeps it under control is a conversation that is happening between your cells, and the language of that conversation is proteins. Proteomics will allow us to listen in on that conversation, and that will lead to much better way to treat cancer.

Introduction
by John Brockman
The leaders of the National Cancer Institute," says Danny Hillis, "are very keenly aware of how little progress has actually been made in the treatment of cancer. This is something they pay a lot of attention to. They're thinking very laterally in giving funding to people like me to work on cancer."
"What they've said is 'Let's bring some new kinds of thinking to this, and create a program where we have physical scientists be the principle investigators, partnered with the co-investigators who are clinicians and biological scientists.' I'm partnering with David Agus, for example. But giving money to the physical scientist is a pretty radical idea, you can imagine it is very controversial within the biological community."
"NCI has started a few of these centers, and given them five years to work. They need to be interdisciplinary and geographically distributed. Our center at USC has people all over the United States involved in it, like Cold Spring Harbor, Stanford, Arizona, UT, NYU and CalTech.
"As a result, Hillis is the newly appointed professor of research medicine at the Keck School of Medicine at the University of Southern California (USC). And he is the principal investigator of a five-year government program on cancer.
The University of Southern California Physical Sciences-Oncology Center's (USC PS-OC) overall goal is to thoroughly understand therapeutic response.
Investigators will establish a predictive model of cancer that they can utilize to determine tumor steady state growth and drug response, particularly those involved in the hematological malignancies of acute myeloid leukemia and non-Hodgkin lymphoma. Furthermore, multi-scale physical measurements will be unified with sophisticated modeling approaches to facilitate the development of a model that can derive the tumor's traits during its growth and after any distress, such as chemotherapeutic treatment. These investigators will apply pioneering measurement platforms to resolve real-time protein interactions and protein abundance and to characterize protein modifications. Appropriately, these studies will also address tumor and host response to therapy using a systems approach. Overall, the predictive tumor response model should enable clinicians to determine the most efficacious therapies a priori and reduce deleterious side effects.
Hillis continues..."We misunderstand cancer by making it a noun. Instead of saying, "My house has water", we say, "My plumbing is leaking." Instead of saying, "I have cancer", we should say, "I am cancering." The truth of the matter is we're probably cancering all the time, and our body is checking it in various ways, so we're not cancering out of control. Probably every house has a few leaky faucets, but it doesn't matter much because there are processes that are mitigating that by draining the leaks. Cancer is probably something like that.
"In order to understand what's actually going on, we have to look at the level of the things that are actually happening, and that level is proteomics. Now that we can actually measure that conversation between the parts, we're going to start building up a model that's a cause-and-effect model: This signal causes this to happen, that causes that to happen. Maybe we will not understand to the level of the molecular mechanism but we can have a kind of cause-and-effect picture of the process. More like we do in sociology or economics.
"Last year's EdgeMaster Class in Los Angeles featured George Church and Craig Venter lecturing on Synthetic Genomics. Hillis points out that the genome is used to construct things, and that it's not the best place for analysis of what's going on. "Certainly," he says, "there are times it is useful, but I don't think that's where most of the information is.
If you think in terms of computer models, think of proteomics as a debugging tool for genomics programs. "When you write a computer program, the first thing you do is you try to run it, and it almost always has a bug in it, so you see what happens, and you debug it, you stop it in the middle of running, and you see what the state of the system is, and you understand what your bug is, and then you change the program. The proteome is the state".
"Proteomics was made possible by genomics. It builds on top of genomics. I guess it's true in some theoretical sense that eventually you might not even bother to look at the genome if you can see the whole proteome, but in practice, it's been very important."
"The genome is the instructions for the cell. That's very important if you want to do manipulation. If you want to actually affect the pathway, then that is the level at which you need to manipulate things. You want to knock out a gene, or modify a gene. Experimentally, being able to read and write the genome is incredibly important. But if you want to use it as a diagnostic for what's going wrong with a particular individual, it will be unusual for that information to be in the genome."

I am talking to Hillis in Villa Miravelle, an historic, exotic mansion with an interesting history, built in 1884 in Saint Helena, CA, by Mexican-American Tiburcio Parrott. In 1974 Spring Mountain Vineyard and winery were established on the surrounding property.

Swiss financier Jacob E. "Jacqui" Safra acquired the Spring Mountain Vineyard in 1991 and through acquisitions expanded the property to 225 acres of vineyard on an 850 acre estate, now the largest vineyard in terms of contiguous acreage in the Napa Valley. Safra, a member of the Edge community, graciously offered the use of Spring Mountain Vineyard as the venue for the Edge Master Class 2010.
This year, to try something different, Edge ran a conference on the East Coast in July (See "The New Science of Morality") There was no intent to run the usual Master Class in California, until Safra came forth with his generous offer, followed by a conversation I had with Danny Hillis a week ago on "proteomics". Given his excitement about the prospects of his new research program coupled by his track record as the man who broke the von Neumann bottleneck to give us the massively parallel computer, I didn't hesitate to announce this event.
The event occurred with one week's notice, the same week as the "Sci-FOO Camp" at Google and the first "Techonomy" conference at Lake Tahoe, both interesting, even exciting events. In the end, among all the usual suspects, it was Hillis, Stewart Brand, and myself who showed up for a weekend at the most exquisite vineyard property in California (See photos below). The weather was beautiful. We were surrounded by dozens of bottles of Safra's prize-winning Spring Mountain Elivette (2005).

Stewart Brand, John Brockman, Daniel Hillis
We a 2-part Edge Master Class which is available below in three formats: streaming viideo (two one-hour talks), audio download, and text (including a printable text file of Parts I & II) .
- JB
W. DANIEL ("DANNY") HILLIS is Chairman and Chief Technology Officer of Applied Minds, a research and development company creating a range of new products and services in software, entertainment, electronics, biotechnology and mechanical design. Hillis is also Judge Widney Professor of Engineering and Medicine of the University of Southern California (USC), professor of research medicine at the Keck School of Medicine, and research professor of engineering at the Viterbi School of Engineering. Previously, Hillis was Vice President, Research and Development at Walt Disney Imagineering, and Disney Fellow. He developed new technologies and business strategies for Disney's theme parks, television, motion pictures, Internet and consumer products businesses.
An inventor, scientist, engineer, author, and visionary, Hillis pioneered the concept of parallel computers that is now the basis for most supercomputers, as well as the RAID disk array technology used to store large databases. He holds over 150 U.S. patents, covering parallel computers, disk arrays, forgery prevention methods, and various electronic and mechanical devices.
As a student at MIT, Hillis began to study the physical limitations of computation and the possibility of building highly parallel computers. This work culminated in 1985 with the design of a massively parallel computer with 64,000 processors. He named it the Connection Machine. During this period at MIT Hillis co-founded Thinking Machines Corp. to produce and market the Connection Machine.
Thinking Machines Corp. was the leading innovator in massive parallel supercomputers and RAID disk arrays. In addition to designing the company's major products, Hillis worked closely with his customers in applying parallel computers to problems in astrophysics, aircraft design, financial analysis, genetics, computer graphics, medical imaging, image understanding, neurobiology, materials science, cryptography and subatomic physics. At Thinking Machines, he built a technical team comprised of scientists and engineers that were widely acknowledged to have been among the best in the industry.
In 2005, Hillis and others from Applied Minds initiated Metaweb Technologies to develop a semantic data storage infrastructure for the Internet, and Freebase, an "open, shared database of the world's knowledge". That company was recently acquired by Google.
Hillis has published scientific papers in journals such as Science, Nature, Modern Biology, Communications of the ACM andInternational Journal of Theoretical Physics and is an editor of several other scientific journals, including Artificial Life, Complexity, Complex Systems, Future Generation Computer Systems andApplied Mathematics. He has also written extensively on technology and its implications for publications such as Newsweek, Wired, Forbes ASAP and Scientific American. He is the author of The Pattern on the Stone. He is a Member of the National Academy of Engineering, a Fellow of the Association of Computing Machinery, a Fellow of the International Leadership Forum, and a Fellow of the American Academy of Arts and Sciences. He is Co-Chair of Long Now Foundation and the designer of a 10,000-year mechanical clock.
We are pleased to present below the entire 2-part Edge Master Class which is available below in three formats: streaming viideo, audio download, and text.
![]()
CANCERING
Listening In On The Body's Proteomic Conversation (Part I)
Right now, I am asking a lot of questions about cancer, but I probably should explain how I got to that point, why somebody who's mostly interested in complexity, and computers, and designing machines, and engineering, should be interested in cancer. I'll tell you a little bit about cancer, but before I tell you about that, I'm going to tell you about proteomics, and before I tell you about proteomics, I want to get you to think about genomics differently because people have heard a lot about genes, and genomics in the last few years, and it's probably given them a misleading idea about what's important, how diseases work, and so on. ...
~~~
Printable text file (Parts I & II)
You've probably heard the genome described as like a blueprint for producing an organism. That's a very misleading analogy because a blueprint is interesting because it says how everything is connected, and how the parts relate to each other. In fact, the genome, at least the part of the genome that we understand how to read, actually doesn't tell you that at all. It's kind of a list of the parts. It does have some control information on it about when different parts should be made, but for the most part we don't know how to read that control information right now. What we know how to read is the parts list. While that's a very useful thing, it's probably not the most important thing that we need to know to understand what's going on.CANCERING: Listening In On The Body's Proteomic Conversation (PART I)
W. Daniel Hillis
Right now, I am asking a lot of questions about cancer, but I probably should explain how I got to that point, why somebody who's mostly interested in complexity, and computers, and designing machines, and engineering, should be interested in cancer. I'll tell you a little bit about what I am doing in cancer, but before I tell you about that, I'm going to tell you about proteomics. Before I tell you about proteomics, I want to get you to think about genomics differently because people have heard a lot about genes and genomics in the last few years, and it's probably given them a misleading idea about what's important, how diseases work, and so on.
Let me start by talking about genes, and giving you a different way of looking at genes, I want to start by clearing up, well maybe not misunderstandings, but putting a different emphasis on how genes work. That will explain why I'm interested in proteomics, and that will explain why I'm interested in cancer.
An analogy might be restaurants. Let's say you were trying to understand the difference between a great restaurant and a bad restaurant, and what you had to work with was a list of the ingredients that they had in their storehouse. Sure enough, if you snuck in at night and looked at their inventory list, you might be able to tell some things about the restaurant. You could probably tell the difference between a French restaurant, and a Chinese restaurant just by the ingredients list. And indeed, you can tell the difference between a European person, and an Oriental person just by looking at their ingredients list, but you probably can't tell a lot about what their personality is like.
Now, sometimes you can tell about defects, so if a restaurant was completely missing salt, or they only had lard for oil, you could say, "Well, this restaurant might be improved if they started using salt, or if they had some butter instead of lard", or something like that. So there might be some gross things that you can tell about inadequacies, that they were missing a key ingredient, something was broken about the ingredients list, but to really understand whether the food was good or not, or how they were making food, you really have to watch what's going on in the kitchen, and watch the process. You have to actually watch the dynamic process; the list of ingredients doesn't work.
The way I think about this is more like computer programs. The genome is like a listing of your operating system, but missing all the control information, so missing all the jumps, and things like that, and that's a fairly useless thing to have if you're trying to figure out, if you're trying to debug a program. It's not totally useless, but it's not that informative. What a programmer would really want to know is they'd like to dynamically look at what's going on inside the machine, what's getting loaded into the registers. That kind of dynamic trace is the much more useful thing for debugging them in kind of a partial listing of the code. If I put it that way, you might ask what's the big deal about the genome, why all the excitement about genomics?
I think it has a couple of historical reasons. One of them is the gene is the great theoretical triumph of biology, it's the one kind of theoretical construct that was predicted. Like the physicists predicted there ought to be a positron, and when they looked, there was a positron. That happens all the time in physics. The equation says there should be a black hole, and we look, and we find black holes. In biology, that almost never happens, and the great dramatic example of it happening was genes. So genes were kind of theoretically predicted by Mendel and they were the core of what Darwin needed, that unit of inheritance. Then Watson and Crick looked and actually discovered it! It was like actually finding the black hole that was predicted. That was in some sense the most exciting thing that ever happened in biology, and since it so stands out, there's nothing close to that, it almost has a religious significance in biology. It is the triumph of the one great theory.
The other thing about it, a practical thing, is that it turned out that people like Kary Mullis worked out this very neat way with tools that the biologists had on their bench, so they could actually measure a gene. In fact, you can almost do genetics in your own kitchen with a few extra pieces of equipment, if you have the right enzymes around. You heat something up, and cool it down, and heat it up, and cool it down, and then you pour it in some jello, run an electric field across it and you actually get a read-out of this nice, digital picture.
So not only was it a theoretical construct that had been predicted by biology, but it was also accessible to experiments with the stuff that people had lying around in their labs. Of course, now we sequence genes with much more sophisticated equipment that does it much more rapidly. But it got its start because everybody could do it in their lab. They could see the genes, so everybody could get in the genetics business immediately, and start getting really interesting genetics results.
For instance, the field of zoology was transformed by being able to tell what's related to what, like kind of the trick of telling the difference between the French restaurant and the Chinese restaurant. By looking at the ingredients you could find the complete tree of family relationships, and so there's a huge amount of good science that suddenly became possible. You could get a lot of hard data. Of course, people immediately looked at what medical applications it could have.
There are dramatic medical examples where you're missing a key ingredient, or one of your ingredients is broken, when there is a disease associated with that - a mutation in a gene, or a missing gene, or duplicated gene or something like that. Cystic fibrosis is an example of that, where the problem is in a single gene. So there are definitely examples like that, conditions that can be identified, and understood in a certain sense by looking at this parts list, or looking at this ingredients list. But if you really want to know what's going on, in most cases a much more interesting thing to do is to look at the dynamics. That is in the proteins that are actually getting generated. Some of them are getting generated directly from genes, some of them are getting generated and then modified by after they are produced. There's a lot that happens after the genetics. And the proteins are controlling which genes are expressed.
So, to me, there is a much more interesting kind of analogy, based on process. The analogy we have so far is about structure. We emphasize the structure of things, so we think of the building blocks, and the things that get built, and the parts. I think it's much more interesting to look at the process that builds all of these parts.
It's true that the human body is an amazing structure, but what's much more interesting is the process that builds it, that maintains it, modifies it. That's not really in the genes, it's in the conversation that's happening between all the parts of the body, and the conversation is happening within the little molecular machines within the cell, or between the cells in the body. Your body has tens of trillions of cells in it, more than the population of the earth, and all these cells are talking to each other, sending each other signals, there's signaling going on within the cell.
To emphasize this other way of looking at it I like to look at the genome, not just as a parts list, but as the vocabulary list for this conversation. It's a useful thing to know, but the really interesting thing to do is to listen in on the conversation. What are these machines all saying to each other? That's what proteomics is about.
Proteomics is the study of all the proteins. "Omics" means "the study of all". The idea became popular when people like Wally Gilbert who started saying, "We should have all the human genes." Then by generalization, people were saying, "Well, we should know all the proteins in the body, we should know all the connections between neurons, and we should know all the metabolites." There are a lot of different kinds of "omics".
What is really interesting about proteomics, is the dynamic conversation; it's the study of the molecules that the genes are making, the ones that are controlling the genome. It is the conversation between the parts. This conversation is happening within the cell, and between cells, the elements of this conversation are proteins that are being sent around, and being absorbed by the cells, or being sent from one part of the cell to another. It's taking place in the medium of proteins, and so if you could see where all of those proteins are, and how they're dynamically changing, then you would, in fact, be listening in on the conversation. That would be a great thing to hear.
Biologists have recognized for a long time that it's a great thing to do. They've tried to do it. It's turned out to be technically much, much more difficult than genomics, for a couple of reasons. One of them is it's essentially an analog process, not a digital process. It matters how much of the protein is there. But another thing is there just wasn't this wonderful technology for dealing with it like replicating DNA.
You couldn't really do it well with the equipment that was lying around in the lab. People have tried to do it, but it was a very unrepeatable process, a very noisy process, and so the first publications about it tended to be wrong because people had mismeasured it, they couldn't measure the same thing a second time. So basically what happened was that it kind of got a bad name in biology, and people said, "Well, we can't get much useful information out of this", because, in fact, they couldn't get it with the stuff they had lying around the lab.
That's where I came in. I had looked at this in the abstract, years and years ago, I thought that this would be a great thing to do, but when I looked in to the details, I thought it would be too difficult. Then just a few years ago, I got approached by the oncologist, David Agus, who said, "We really need this information for treating cancer patients", and he convinced me to look at it again with the new tools that had come along.
The tools typically are things like mass spectrometers for weighing molecules, and liquid chromatography, which is basically sliding a molecule past a bunch of other molecules, and seeing how much it sticks. We can also make antibodies that stick to very specific molecules. That is a set of tools that hasn't changed very much, but when I started looking at it, I realized that the big problem was that people were using these tools basically in a lab bench, and treating it almost like they were treating genomics, as if it was a digital process. They were going through a sequence of experimental steps, but the way that they were controlling it wasn't possibly good enough to even get the same result twice if they measured the same sample, much less to look for subtle things in the changes.
I realized that it really needed a couple of other things, one of which is some better application of physics, which is how the instruments were actually tuned to do this problem. Another thing it needed was some plain process engineering. What was required was much more like making a semiconductor line than it was like sequencing DNA. There were many, many steps that had to be refined, and highly, highly controlled in order to get a repeatable result in protein. So this was essentially an engineering problem.
There are certainly hundreds of thousands of different protein variants, and maybe more. Nobody really knows which variations are significant. But certainly every gene produces a protein, and then those proteins get modified by the processes, and combined, and produce other proteins, and so on. The big problem was that there was no way of looking at all the proteins, say in a drop of blood that was repeatable, that you could measure the same drop of blood and see the same proteins, and part of the problem is because they occur in vastly different amounts. Some of them are a million times more diluted than others, so there's a huge dynamic range.
But also there are hundreds of steps in the process of measuring them. So if you're doing this with graduate students in a bio lab, and one of them goes and has a cup of coffee at one step, and you leave it 15 seconds longer than an enzyme, you get a completely different result. What needs to be done is a super tightly controlled engineering process.
Since that was essentially an engineering problem, I thought that could be an interesting problem for an engineer like me to work on, so I started working on that. Then it turns out that once you get that, there's a huge mathematical problem at the end, which I was also interested in. It was a computing problem, of interpreting all of these results. If you know that this protein is going up or down, how do you make any sense of that, and correlate that to anything useful. That is essentially a computing problem. Since it was a computing problem, and an engineering problem, I thought that I had something to bring to the table, and started working in the area really just to get the engineering worked out.
Applied Minds can do projects in the exploratory stage without going off and getting any funding; we do it with our own profits from other projects, so we started exploring proteomics with David Agus. And that's how we realized that we could do it if we could really build a line like an assembly line for doing it, which involved robotics, and changing the mass spectrometers, and things like that.
We got to the point where we actually knew how to do it, and at that point we raised some money from some angel funders, and made a company called Applied Proteomics, which has worked out how to do this, and built this assembly line which does these hundreds of steps, and measures along the way, and does it in an automated way. For the first time, the results are accurate and repeatable. When we test the same sample, we get the same result.
You can take a drop of blood, and get a repeatable measurement over a hundred thousand repeatable stable features. We don't know necessarily what all of them are, but many thousands of them we can identify as known proteins, and we now have genes associated with them. Often that means we know something about the function, or where they are created in the body, or something like that.
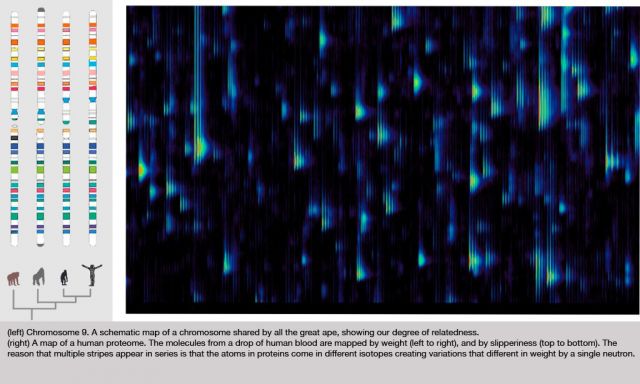
Let me show you the results of that process, which is on this slide.
Figure 1: Differential Feature
This is actually a small part of the measure that we get out of a drop of blood; this is actually a small part of a bigger picture. We've spread out the fragments of proteins in two dimensions here. It's a little bit analogous to a gel that you might see, or a gene chip, the same protein feature will always appear in the same position every time. The brightness shows how much of that protein there is. This display actually doesn't show you too much of the dynamic range and brightness, but we are measuring that.
In the horizontal direction we're measuring the mass of the protein fragment. The vertical axis is how slippery it is. People have produced pictures like this before, but what's interesting here is that every time you do this, the features come out in exactly the same places. That hasn't been true before.
Just to show you how precise these pictures are, you notice that these things tend to occur in these little groups of stripes, tick, tick, tick. You see there are several of them in each group, and they kind of trail off; it's almost like a ring, or an echo. Well, the reason for that is that carbon has different isotopes, and so if there is an extra neutron, you have a different isotope of carbon in the protein, then it's going to be slightly heavier. That distance between the stripes is actually the weight of one neutron; it gives you an idea of how precisely we're measuring things. There's nothing in between because there's no such thing as half a neutron. In fact, measuring things so precisely we can often tell by the shape, how many carbon atoms there are in the protein.
Figure 2: Yellow Overlay
The amazing thing about this picture is I'm actually showing you two measurements of two different blood samples on top of each other. One of them is shown in red, and one of them is in green. Things look yellow because the two are exactly on top each other, because the two blood samples are mostly the same. But, if I look closely at this ... actually let me just find a spot which is different ... okay, well, actually here is a good spot. There is some protein that was in one of the blood samples that wasn't in the other one, so you only see it as the green. There is another place where you see there's a little red down there. That is something that is in one sample and not the other.
It's almost as though we've got a digital read-out of this highly analog process. That's the amazing sort of engineering feat that I don't believe anyone else has achieved that kind of repeatable precision over that much range before. What we know is the relative concentration of each of those proteins. Now, this pair of tests might be the same person at two different times, or it might be two different people. They probably both have the gene to produce that protein, but for some reason one of them is saying this, and the other one isn't saying this at this time.
Now if we have a hundred thousand of these features — and we do have more than a hundred thousand — then the question goes on to what do they mean, what do we do with it? That's the stage that we're at now. It may be that some of those like a genetic test, maybe a single feature will actually tell us something. But probably much more of the information is in the patterns and combinations, and so on.
For instance, let's say that we go to cancer patients, and we try out a drug on them, and we find out that only 10 percent of them respond to the drug. It would be very nice, if there were some genetic marker that told us which 10 percent responded to the drug, because it's a miracle drug for those 10 percent, but it's a useless drug if only 10 percent of the people respond to it, and it makes 20 percent of the people sick. You would like to know which are which, and it was a great hope that maybe you would be able to find genetic markers to do that. There are a few drugs that that's true for, but by and large, that information doesn't seem to be just in the vocabulary list.
But the information is much more likely to be in here, there's something dynamically happening, and so if we can start to say, "If we see this pattern approaching expression, it means that you've got this thing going on metabolically." All of a sudden we've got hundreds of thousands of symptoms to look at, if you will, or hundreds of thousands of indicators of what is going on at the level of what's actually going on.
In the process, I started looking at how we treat cancer, and how we think about cancer. This is another area where I think there's a wrong paradigm that has gotten started because there's been a great success, and that success has been over generalized. In this case the great success has been the treatment infectious diseases and the germ theory of disease. This is the greatest success of a theory in medicine.
That was a very cool development, because if you can figure out what species of germ you were infected with, then that sets how you should treat the disease. You could treat the disease with something that would kill that germ. That became the general paradigm of medicine. You would do a diagnosis, a differential diagnosis to figure out what the infectious agent was, and then you would apply a treatment that was very specific for that agent.
That's the thing that doctors are basically trying to do, identify the disease, and treat the diagnosis according to the best method. That allows science to come in because you can objectively test whether a particular treatment is effective, or not effective, when dealing with that diagnosis. Does quinine help the symptoms of malaria? Is penicillin the best way to treat anthrax? Once you know what's best, that's the thing that doctors are taught to do.
Interestingly enough, that way of looking at things is not the only one in the history of medicine. Historically, doctors had theories that are today more like Ayurvedic medicine, with its emphasis on balances between various forces in the body. Or in the West, a medieval doctor might have tried to make you less choleric or more phlegmatic. The idea was to try to restore the order of the various forces that were controlling the body. It's interesting, at the time that the germ theory of disease was really exploding, and antibiotics were being discovered, J.B.S. Haldane said, "This is a disaster for medicine because we're going to get focused on these germs, and we're going to forget about the system." He was right.
Indeed, if you look at what happened, it was a disaster for treating diseases like cancer because we started thinking of them almost like they're infectious diseases. It's a habit of thought, so when a patient comes in, we diagnose them, and we put them in a category, and then we try to apply the treatment that is shown to work on that category. We do a blind clinical trial of how people that are in that category respond to a certain drug. That makes a lot of sense for infectious diseases because infections are species, they speciate, and divide out, so putting them in categories makes a huge amount of sense.
But a systems disease like cancer, or an auto-immune disease, is a break down in the system, much more like a program bug. We would never think of debugging a computer by putting it into one of twelve categories, and doing something based on the category. Actually we do, it is kind of "help-desk debugging" that doesn't work very well in complex situations.
There is a big difference between help-desk programming debugging, and the kind of debugging a programmer really does when they're trying to more subtly fix a program.
What we've got in medicine now is kind of help-desk debugging. We put you into a category. In cancer we start by putting it in a category that's based on the part of the body where symptoms of the cancer have been shown. Then we test drugs that way: Does this drug work on lung cancer, and if it does, well, it's not approved for prostate cancer because we tested it on lung cancer. That's a whole other experiment, that's a different category of disease. Then we subcategorize them. We take a biopsy sample, and we say, "Well, these cells are kind of squishy and long, and those are kind of round, so we have the squishy, long cancer, and the round cancer." We declare that we have two forms of breast cancer.
We keep coming up with more kinds of cancer as we measure more things, and then we subdivide the categories. There used to be dozens of kinds of cancer, and now there are hundreds of kinds of cancer. But I actually think there are millions or billions of kinds of cancer. Cancer is a failure of the system. Happy families are all alike, but unhappy families are all unhappy in their own special way, and happy bodies are kind of all alike, but when they break down, they all break down in their own special ways.
The breaking down is at the level of this conversation that's going on between the cells, that somehow the cells are deciding to divide when they shouldn't, not telling each other to die, or telling each other to make blood vessels when they shouldn't, or telling each other lies. Somehow all the regulation that is supposed to happen in this conversation is broken. Cancer is a symptom of that being broken, and so when we see a whole bunch of cells starting to divide uncontrollably in an area, we call that "cancer", and depending on the area, we'll call it "lung cancer", or "brain cancer". But that's not actually what's wrong, that's a symptom of what's wrong.
To use another kind of analogy, let's say we didn't understand anything about plumbing, but occasionally we came home and our living room is filling up with water, and sometimes we come home, the kitchen is filling up with water, and so we start describing the problem as, "Well, my house has water, that's the problem." We might even divide it and say, "My house has kitchen water, or my house has living room water." If plumbers were like doctors the best they might be able to say is "we've learned about kitchen water, and if we pour a lot of drano in the kitchen, then kitchen water sometimes goes away. Living room water is fixed by pouring a lot of tar on the roof." Indeed, there might be ways of fixing the problem, but what you really need is to understand about plumbing. You should be worried about the process that's creating the water, and understanding about what's supposed to be draining, and what's supposed to be holding it, and so on.
In fact, we misunderstand cancer by making it a noun. Instead of saying, "You know, my house has water", we say, "My plumbing is leaking." Instead of saying, "Somebody has cancer", we should say, "They're cancering." The truth of the matter is we're probably cancering all the time, and our body is checking it in various ways, so we're not cancering out of control. Probably every house has a few leaky faucets, but it doesn't matter much because there are processes that are mitigating that, by draining away the leaks. Cancer is probably something like that.
In order to understand what's actually going on, we have to look at the level of the things that are actually happening, and that level is proteomics. Now that we can actually measure that conversation between the parts, then we're going to start building up a model that's a cause-and-effect model: This signal causes this to happen, that causes that to happen. Maybe we will not understand to the level of the molecular mechanism but we can have a kind of cause-and-effect picture of the process. More like we do in sociology or economics."
Whatever the treatment of cancer, or auto-immune disease, neurodegenerative disease or other system diseases will be like in the future, there won't be a diagnosis step, or at least that's not what will determine your treatment. Instead, what we'll do is we'll go in, we'll measure you by imaging techniques, and taking it off of your blood, looking at the proteins, things like that, build a model of your state, have a model of how your state progresses, and we'll do it more like global climate modeling.
We'll build a model of you just like we build a climate model of the globe, and it will be a multi-scale, multi-level model. Just as a global climate model has models of the oceans, and the clouds, the CO2 emissions, and the uptake of plants and things like that, this model will have models of lots of complicated processes happening at lots of different scales, and the state variables of this model will be by and large the proteins that are moving back and forth, sending the signals between these things.
There will be other things, too. But most of the information is in the proteins. There will be a dynamic time model of how these things are signaling each other, and what's being up-regulated, and down-regulated, and so on. Then, we will actually simulate that under lots of different treatment scenarios; we'll simulate for your cancering, how we can tweak it back into a healthy state, having it guided back toward a healthy state. It will be a treatment that's very specific. We'll look at those and see which ones are most likely to bring you to a healthy state, and we'll start doing that, and we may treat you in a very different way than we've ever treated any other human before, but the model will say that for you that's the correct sequence to treat it.
Right now this would be a huge change in medicine. For instance, the way we pay for medicine is dependent on the diagnosis. You pay a certain amount for prostate cancer, and you pay a different amount for lung cancer. That determines what part of the hospital you get routed to, which doctor sees you, what the insurance company will pay for. If you take that out of the system right now, it's a completely different kind of a system. I don't think this will be an easy switch and I don't know what the sociological/economic processes will be. But it will happen because it will start working better.
It will probably happen first with desperate people who aren't getting fixed with the normal methods, will go to this alternate process, and when enough of them start getting fixed by this alternate process, then that will, by some complex sequence that I won't even try to predict, eventually change medicine.
Of course there is a lot to be done to make this work. We are dealing with very different time scales and different space scales, too. There are useful things that are hour-to-hour time scale, in your bloodstream, and your cell level is probably more like minute-to-minute, or even faster. Right now we're just beginning to be able to measure proteomics within a single cell, and so right now what we are doing with the National Cancer Institute is trying to bring all of those time scales, and space scales together into a model. This is probably, with today's technology, a ridiculous stretch, but we're at least attempting to do it.
We're measuring things both at the inside the cell level, measuring at gene expression the production of proteins, the placement of proteins within the cell, then the conversation between the cells. That we're trying to do at more the minutes time scale, and then we're doing what's happening in the body and the blood, on the days time scale. We should be probably measuring it over hours, but with current technology we can't afford to do that.
We're doing this in mice right now, but we can only draw so much blood from a mouse. Those kinds of things are limiting us. We're also doing it with imaging, the actual geometry of the tumors, so we're actually trying to measure geometry; we're trying to measure the genetic evolution of the tumor because the tumor is not homogeneous. Genetically it's different inside than outside, and we are trying to make a model, like one of those kind of global climate models, for lymphoma in mice. We can get genetically identical mice, and we can actually very reliably give them the same kind of lymphoma, and so we can repeat the experiments.
So it's much better than the global climate models situation. With global climate, we have one experiment to calibrate our model, and we're in the middle of it. There is no control. In the mice we can do a lot, we can try different variables, and then also try what are the effect of different things that we can do in terms of treatments, of giving chemotherapy of various sorts, or heating them up, changing the pH of their blood, doing all kinds of things like that, and begin to get a perturbation model of not just how the system normally works, but how it works under different kinds of perturbations. Then hopefully, eventually, we'll get to the point where we have a good enough model that we can actually predict: if we do this to this mouse we can actually make it live longer.
We are already learning a lot. One is, for this mouse study, we're combining some new techniques like proteomic techniques with a lot of techniques that were developed for other reasons, for instance imaging techniques that are very detailed. We're actually putting little windows into the mouse, and watching the tumor grow, and then we can use things that have antibodies that bind to certain kinds of proteins that are being expressed, and so we can actually see where those proteins are being expressed in the living mouse, so see geometrically where they're being expressed.
There are techniques, for instance, where we can actually look within a cell, and see where a protein is within the cell. We can actually do microscopy below the wavelength of light now, which is a fantastic advance, by using basically little flashes of light, and computing on top of it. There are huge advances in the technique and instrumentation and so on that's making this at least conceivable for the first time. It's only a matter of time before it will be possible, and it's quite probable that this first attempt is too early, but we are attempting to do this with the consortium of people in places like Stanford, and Cold Spring Harbor, and USC, UT, NYU and Caltech. The National Cancer Institute, has actually given our group five years of funding, assuming we keep making progress.
They had this crazy idea of getting people like me, who are not really biologists, to be the principal investigators of these centers, to work with clinicians to design the program of research, which is then being carried out by a lot of people who know things like how to put windows into mice, and how to image a tumor, or how to get antibodies to glow. So we're using all of those biological lab techniques to do something that's really more like a physical sciences model.
I'm optimistic that we'll have enough success that people will at least try to repeat this form of experiment. Whether we actually are able to make accurate predictions is yet to be seen. The group coup would be if we got to the point where we could say, "We can predict that if we do "this" to this mouse, then we can take care of "that" in the mouse". That would be success. But we can learn a lot without getting that far.
![]()
CANCERING: Listening In On The Body's Proteomic Conversation (PART II)
W. Daniel Hillis
What I've been talking about here is more analysis than construction. The genome is used to construct things, and I'm claiming it's not the best place for analysis of what's going on. Certainly there are times it is useful, but I don't think that's where most of the information is. In fact, in some sense, it is literally true that the information that's in proteomics tells you everything that was in the genome, everything useful that was in the genome. In a sense, the genome is redundant if you have the proteomics, that's theoretical though, because the genome is digital, and we actually have it. In many ways it's enabled proteomics. ...
Printable text file (Parts I & II)
(During this session Hillis was asked to comment on a number of specific topics:) CANCERING: Listening In On The Body's Proteomic Conversation (PART II)
W. Daniel Hillis
On the relationship between genomics and proteomic testing
What I've been talking about here is more analysis than construction. The genome is used to construct things, and I'm claiming it's not the best place for analysis of what's going on. Certainly there are times it is useful, but I don't think that's where most of the information is. In fact, it is literally true that the information that's in proteomics tells you everything that was in the genome, everything that was useful. In a sense, the genome is redundant if you have the proteomics. That's theoretical though, because the genome is digital, and we actually have it. In many ways it's enabled proteomics.
Right now, when I show you that image that has the hundreds of thousands of dots on it, I can actually tell you what a lot of those things are because we know the genome; so I can actually associate many of those dots with genes, and because we have these great genetic expression tools and so on, we may know what part of the body it's in, we may know a lot about the pathways because we can do knock-out genes. It's a great experimental method for actually controlling what proteins get produced and so on. In many ways proteomics was made possible by genomics. It builds on top of genomics. I guess it's true in some theoretical sense that eventually you might not even bother to look at the genome if you can see the whole proteome. In practice, it's been very important.
The genome is the instructions for the cell. That's very important if you want to do manipulation. If you want to actually affect the pathway, then that is the level at which you need to manipulate things. You want to knock out a gene, or modify a gene. Experimentally, being able to read and write the genome is incredibly important. But if you want to use it as a diagnostic for what's going wrong with a particular individual, it will be unusual for that information to be in the genome.
On the role of gene testing in cancer
Let's take somebody who has cancer. They used to be somebody who didn't have cancer, and they had the same genome. So the difference between having cancer and not having cancer is clearly not just in the genome. There's more to it than that. In fact, most of their cells aren't cancering, and they have the same genome. Cancer is a dynamic process that's happening, and it's not just in the genome. Now, there may be a specific mutation from a genome that helps explain why it happened. For instance, one of the dramatic genetic test successes has been in breast cancer, BRCA1 and 2, which are specific genes that are associated with breast cancer. They occur a lot in Ashkenazi Jews, and a particular kind of breast cancer is associated with these genes. There are many examples like this where there's a genetic predisposition to cancer, but is one of the clearest examples. The cancer isn't inherited, but that's a predisposition that is, so people that have the gene are more likely to have cancer.
Let me put it in terms of the conversational analogy. That means that certain words are missing, and in that case we know there is a conversation about fixing broken DNA. We're repairing broken DNA and it's hard to describe what to do without those words. You need to discuss BRCA1 and BRCA2, and you need to use those concepts. You need to use those words in order to repair DNA in a certain way. If you don't have those words in your vocabulary, then you're unable to execute this process of repairing DNA.
If those words are slightly mutated so that they're not understandable, if you slur them, or stutter them or something like that, they're won't have the desired effect. It won't always cause a problem because there may be other pathways that also repair the same defect. If the other pathways are working very well to control your DNA, it might not matter. But there is an association, and you can look at people, you can test them, and you can say, "Well, if they have this mutation, they're more likely to get breast cancer", and actually, it turns out, they are much more likely to get breast cancer.
Furthermore, you know what the failure mechanism is, so there are many people who get breast cancer for completely different reasons that have perfectly intact BRCA1 and BRCA2. By the way, that same pathway, because it's a general pathway for repairing DNA, is also important for ovarian cancer because if you don't repair properly, then you're more likely to get ovarian cancer, too. It doesn't really have anything much to do with breasts other than that's where you see the symptoms of this happening, it's usually first noticed in breasts.
On the application of proteomic modeling to cancer treatment
The cancering metaphor does mess up our standard model of medicine, where we just take the right pill to fix a given problem. But if you think about it, the idea that you should be able to take a pill, and it should magically fix a disease, a systems disease, a failure of the system, is kind of amazing that that's even possible. The cases where it's mostly possible is where you have an invading thing that doesn't belong, like an infectious disease, and you take a pill that poisons that particular thing, like an antibiotic. There are a few cases where you're just missing one component, and you take a pill that provides the missing ingredient, and so there will be a few magic cases like that, but those are very special kinds of failure, and I don't think they'll be the typical failure in cancer.
Unfortunately this whole idea of fixing a disease with a pill, while it's delightful when it works, is not very generalizable. We haven't found very many new pills lately that really cure diseases. In fact, the pharmaceutical industry is kind of broken right now because they've run out of this low-hanging fruit, a magical chemical that cures a disease. I don't think we're likely to find a lot of more those. They need a different model.
The first commercial application of proteomics will be diagnostics and probably something much simpler than what I was describing before. The way proteomics will get started is by being markers for diseases that we already diagnosed, in other words, it will help support this categorization system of diagnosis.
Let's say I could find a pattern of proteins expressed in your blood that said whether your colon was growing polyps in it. That would be much better than having a colonoscopy. You get a colonoscopy every five or ten years, which is now recommended — even though in one in a thousand people they create serious damage giving the colonoscopy — we still recommend that people do it because we don't have a better way of telling if you're in this kind of precancerous stage. If you could do that in your annual blood test, it would be much better. That is an example of a very early use of proteomics. Probably there would be many things like that where if you could detect breast cancer without a mammogram, or if you could do a confirmatory test of prostate cancer without doing a needle biopsy, all of these things are very invasive, and expensive, and cause a lot of secondary harm to people.
On the relationship between proteomics and synthetic genomics
At last year's Edge Master Class in Los Angeles, George Church and Craig Venter talked about Synthetic Genomics. Proteomics is relevant to that because it's also a tool that such researchers could use since they need to debug their synthetic genomics. When you write a computer program, the first thing you do is you try to run it, and it almost always has a bug in it, so you see what happens, and you debug it, you stop it in the middle of running, and you see what the state of the system is, and you understand what your bug is, and then you change the program.
Right now George and Craig don't have the debugger. Proteomics is the debugger that they need. When they write a program and it doesn't work, which actually happens a lot, in order to tell why it's not working, they need proteomics to say, "Oh, I see, this isn't upregulating that enough, or downregulating that ... ", and that will help them debug their program and tune their program. Right now a lot of synthetic genomics is about copying a naturally-evolved program, and saying, "Okay, I can make a copy of this program, and write it, and it does the same thing", and that's interesting. It would have been very surprising if it hadn't worked.
On the business of proteomics
The business of proteomics had a false start a few years ago. As I said, proteomics can't be done with the techniques and tools that are sitting around a laboratory, a biological laboratory; it's not a biology problem. Unfortunately there are a lot of people who tried to do it with those tools and so there were a few companies that started up, and a lot of laboratory projects that started up, and they published a bunch of results probably prematurely, and they couldn't replicate them because, you know, the next time they ran the tests it came out differently. It was so noisy that they had to do very, very large trials. So if you have a bad instrument, then it's not going to work.
Because of that, proteomics got a bad name among the venture capitalists. Probably most venture capitalists will cut and run if you say "proteomics" right now because of the problems with the tools of a few years ago. What will happen is we'll get some successes. In spite of this, there will be a few things like Applied Proteomics that get started, and there will be a few people with more vision that look at it more closely, and say, "This actually fixes the problem that they had before, because the story was right before, it was just they couldn't make it work." As soon as there is a success, then you'll see a general change in attitude. Venture capital tends to work this way. There will be lots of people investing in this area, and there will be a boom very much like there was in genomics, and sequencing. Lots of effort will go into both the technology of doing the proteomics better, as it happened with genomics, and also the application of proteomics.
And again, first it will be to these diagnostics, and then it will be things like drug rescue. Billion-dollar drugs that had to be taken off the market after having had huge amounts of money invested in them because one in ten thousand people reacted badly to them. Well, if you could tell who was going to have that reaction, if you could make a test for it, and again, if you could find in a proteomic marker for what it was about, what was wrong with the dynamics of their body to cause them to have that reaction, then all of a sudden such drugs would be viable again. They could recover that billion-dollar investment. You'll see pharmaceutical companies take it up for reasons like that. That's the "safe" part pharma companies are looking for drugs that are "safe and effective", so it will help them with safe.
It will also help them with "effective" because right now, if you have something that not everybody responds to then it is not effective. We've had the first examples of that already, where a drug wasn't statistically effective. But when we look again we can say, "Well, people who have this protein being expressed, it is effective." So there is a very simple case where you're looking at a single protein. This single-protein expression is a situation analogous to a conversation with somebody shouting "Fire". A single protein is a very degenerate case of the conversation. There will be a few markers like that, that are simple markers, but mostly I think we'll look at things that are much more complex patterns that happen in multiple proteins.
Dosage is another reason why pharma companies will get very interested in proteomics, and it probably will make a big difference in research. Right now you can't tell what's happening until it gets all the way up to having a symptom in a patient. If you have something that takes a long time to play out, like Alzheimer's disease, or ALS, you really don't know if your drug is doing any good for years and years. You have no idea if the dosage is too high, or the dosage is correct. The feedback loop is very, very indirect, and lots of other things are affecting it, too. But if you could actually look at the proteins and notice this bad problem of communication between the cells, that's causing plaque formation in the brain in the case of Alzheimer's, then you might be able to see the response of the drug immediately, even though the symptoms aren't changing yet. The symptoms may take years to change, but you can see the drug is effective in this patient immediately… or that it is not, so you can move on to trying the next drug.
You could titrate the dose and very quickly say. You can titrate the dose not only so the guessing holds the correct, safe dose, but actually specifically for the patient. We know that people respond very differently to drugs. Right now it's a trial and error process. They give you a little bit, if that doesn't work they give you more. We see this happening even with simple drugs like blood pressure drugs. Now, with blood pressure, doctors can measure blood pressure easily. They should give you a small dose of the drug first, they see if your blood pressure went down enough, and they can give you a little bit bigger dose. You can do that as a quick loop and you calibrate the response for an individual because you can measure blood pressure. Well, you can't do that for something where the outcome is years away. Proteomics ought to let us do that, too.
On the way proteomics will change treatment
Something that I skipped over in describing the new treatment paradigm, and the simulation paradigm of treating rather than the diagnosis paradigm, is that right now what you do is you diagnose, and you select a course of treatment. Now, what really happens if it doesn't work, maybe you switch and jump on to a different track. But the interesting thing, once you can kind of measure the dynamic state variables, is you're constantly redoing that. You replan every time you go into the doctor. There is not a notion of staying the course, there's no reason to stay the course because you can tell you're getting closer. You redesign a new treatment every time you remeasure. You're constantly getting feedback. You don't just design a special customized treatment per person, you do it per person per time, per person per visit to the doctor.
What you're trying to do is guide it back to a healthy state, basically. But the other great thing is because you're looking at the whole state, you're not just treating one thing at once. That's the other thing that's a flaw in this idea we've gotten about medicine, which is in the infectious disease model. The fact that you have malaria is the main event. You've got to kill off that malaria before anything else can happen. But in the systems diseases, there's lots going on in the body, constantly. So when we say a treatment is safe and effective, the way that we test that is we're testing one thing at once, not looking at everything else. So we say, "Okay, well, some statins lower your cholesterol, which we think is a surrogate for you're going to have a heart attack", so we give everybody a statin.
Now, if you look at some of the early monkey studies where they first studied statins, and proved that it lowered cholesterol, the monkeys that were taking the statin had a higher death rate than the monkeys that weren't taking it. What does that mean? Well, they kind of ignored it, because they looked at the deaths and they were from accidents, or getting into fights, things like that. I bet that it's going to turn out that statins actually cause you to get into fights, and have accidents. It will have effects on your mind that weren't studied in all the studies of statins, and in fact, statins may not be good for you. Or they may be good for you for reasons that have nothing to do with lowering cholesterol.
That's another thing that we're discovering in statins. The idea that we go in there, and we have this very complicated system, and pick one variable and say, "Does this one variable get better when we give this person this pill?", and then say, "Okay, well, it did." The engine rate went up because we poured this goop into the engine. But is that really good? Well, maybe the speed went up because it broke the regulator or because it clogged up the safety valve. So right now when we test the drug, we're looking at one thing at once. We only discover these bad side effects in these retrospective studies looking back after people have been taking the drug for a long time. ?
What most of these drugs do is shift the balance. They're trade-offs. There is a reason why you don't naturally produce statins. I don't think it's because nature didn't think of the idea, it's probably because statins have some pluses and minuses, and so what you're doing is you're rebalancing things, tipping things in a certain direction. The reason that's relevant is if you're in the paradigm of diagnose the disease, treat the disease, then that tends toward treating the disease at the cost of making everything else a little worse, than if you're looking at the whole system, and trying to optimize the whole system. Proteomics is looking much more at the whole system.
Proteomics can also tell you things that can give some substance to what nutritionists have been talking about. If you look at traditional Chinese medicine, or Ayurvedic medicine and things like that, it's all in terms of balances, restoring this force against that force, but it doesn't have a very good model. It is a highly oversimplified model that's been made up without much scientific information going on, just a lot of time going into it. If you had a much better model, then you could probably rationally understand what foods you should really eat that bring your body back into balance.
It is probably very different for different people, and so I don't think the treatment is necessarily going to be taking a pill. You will take pills, but you'll also change diets, or you may discover that it's very important for you not to be stressed out in the afternoon, you may find it's important for you to get lots of oxygen, or do something very aerobic. You may find out that you better not get cold. Or we may deliberately heat you up to an abnormal amount for a little while. Maybe you will need sequences of these things. I don't think treatments will be "a thing", like a pill, I think treatments will be perturbations, bringing you toward health, with some feedback as to what results all those perturbations are having.
In a sense we already have home proteomic tests. Some of these home pregnancy tests may be that. What's happened is, for various reasons, we have luckily discovered certain proteins that are diagnostic, PSA is a great example.
PSA is a protein, a prostate-specific antigen, and for various reasons we happened to notice that this was a marker for prostate cancer, that it was associated with prostate cancer. Now when you go get a blood test, if you're male you probably automatically get a PSA test. That is a protein test — a test for a specific protein. Once you can look at the whole protein, one of the things that will happen early is you'll identify specific proteins to test for, and those may be home tests. They may be finger prick tests, or there may even be indicators of a protein that are excreted in your urine, so it wouldn't surprise me at all if you get home tests that come out that were identified by looking at the whole proteome. But my guess is, again, those are going to be kind of unusual, special cases.
In some sense it's really lucky that there happens to be a single protein that's associated with inflammation in the prostate. In fact, maybe it's lucky, maybe it's not, because there are a lot of people getting needles stuck in them unnecessarily because of this test, and probably they would be much happier not being told they have prostate cancer, because it probably won't hurt them anyway. Maybe the needles are worse than what would have happened without the test. It causes a lot of worry, and stress, and maybe the stress even makes them prone to some other kind of cancer.
Probably there is a much more sophisticated combination of lots of different proteins that you can look at and say, "This is an inflammation of the prostate that's actually not cancer." Or, "This is an inflammation that looks like this particular kind of cancer that responds to this drug, while this is a particular kind of cancer that grows very slowly, and it's not going to kill you. You'll die of something else first, so you don't need to worry about it." Once we get much more subtle measures, once we can listen to that conversation and find out what's actually going on, then I think there will be a lot less treatment. We do a lot of overtreating of things, and a lot of damage by treatments. That's actually one of the only things that can actually reduce the cost of medical care. The cost of medical care is mostly way down the line when people at hospitals, are already pretty damaged by disease and treatment by the time they become very expensive.
If what you can do is spend more of the money up front diagnosing things, tweaking things before it gets to that stage, then that is a win-win because it's better for the patient to get less treatment, and better for the insurance company because you never get to the hospital. Of course, the doctors actually are happy because they really do want to cure the patient. They have more tools to actually know what's going on. It's very frustrating for the doctors to know that a treatment might not work.
Doctors hate this thing of giving you a sequence of poisons: hopefully we'll kill the cancer faster than it will kill you. That's a horrible thing to do to a patient. I think oncologists would love to have better ideas of what the effects of the treatments that they're giving are, and is it actually doing any good. They want to avoid giving people treatments that are not going to help, that are going to make them sick.
On home testing
I'm sure we'll have examples where people can do self-testing, perhaps for a single protein. I think it's going to be rare cases. Sometime we may see proteins or parts of them in the urine. Mostly proteins get broken down, unless you have something like bladder cancer.
Usually we test the blood. But, diabetics do blood draws all the time at home, and tests. So I can certainly imagine things that are the equivalent of diabetic sugar meters, glucose meters that you're adjusting your own dose of blood pressure medication, or anti-cancer medication, or something like that. Right now proteomics makes it very expensive to do because we're just able to do it for the first time. But certainly once you've identified what you're looking for, then there is some fairly standard methods of changing that, and turning that into tests that are very easy. The classical method is you develop an antibody to the protein, and then you can make something that changes color. More and more it will be changes to the conductivity of a transistor so that you won't read out a color, you'll have a little machine that does the analysis, not just indicating presence or absence.
I think the interesting information will be in the levels of many different proteins. The picture I showed you was a protein that was produced in one and not in the other. Much more common, though, we see proteins that change in levels by 20 percent or something like that. A lot of the information isn't just on/off, it's in the concentration. You don't totally turn it on or off, you change the rate at which you're producing it and a lot of drugs basically do that, or they change the rate of production or destruction of something. They upregulate something, downregulate something else. If you take BRCA as an example, there's another protein that I think regulates a promoter of BRCA production, and actually if you have a mutation in that, too, then the mutation in BRCA is less important because it's not regulated. That's a completely different kind of subcategory of breast cancer.
When you try to downregulate something, it is because you have too much of it, so you lower it by giving a drug that suppresses its production or speeds up its destruction It's all in the quantities, it's not just present or absent like it is in the genome, and that's the thing that's really neat about the genome is it really is digital. You either have a gene, or you don't. You have this variant of the gene or you don't. You have one or two copies of it.
I'm emphasizing blood because it's nice you have a collecting system built into your body that goes around and touches every place in your body and collects fluids. It's very convenient for diagnostics. But there will probably be things that happen in tears, or in saliva, or in lymphatic fluid, or spinal fluid, but your body is plumbed very nicely for blood so that you can get it out easily, and since it delivers nutrients, and gets rid of waste, it pretty much is involved in anything very dramatic that's happening in the body. It's good low-hanging fruit, at least.
In diabetes treatment, what we do now is we basically get around your pancreas. Doing what your pancreas does by adding insulin. We try to mimic the pancreas. Proteomics may allow us to actually stop the processes that cause your pancreas to go bad earlier. We know that that's something that can be affected by diet. Again, it's probably predispositions, so if you're missing certain vocabulary words, it's harder for certain kinds of processes to work, and so you are particularly susceptible to certain kinds of problems, so that's where genetics comes out. Then proteomics can see, are you actually having those problems that you're susceptible to, or are your other redundant mechanisms taking up the slack for it? The body seems to be highly redundant, and often has lots of mechanisms for doing anything important.
On redundancy and treatment
Probably the biggest surprise that happened with genetics is once we had the ability to knock out genes, everyone thought, "This will be great, we'll knock out the gene and see what breaks." Well, it turns out mostly when you knock out a gene, at least half the time nothing seems to break. There was some redundant thing that just took up the slack. Probably if you did very careful studies on those knock-out mice you would find out that they were more susceptible to certain kinds of diseases or breakdowns because they're missing one of their redundant mechanisms.
It's interesting that evolved systems, much more than engineered systems, are evolved to be robust and redundant, and if you think about it, robustness is a kind of information hiding. What you wanted to do is you wanted to respond the same under many different kinds of circumstances, and with some of its parts broken. What you're doing is you're hiding the information in effect about, how it's working. The very process of making something robust is a process of hiding the information at the level of the symptoms, at the level the physician is looking at. In a very real sense, evolution has evolved your body to make the systems uninformative about what's going on inside.
It's kind of amazing that a doctor can look at things on the outside of you, and take your temperature, that it works at all that you can look at the outside of somebody and decide what you should do to them to make them better because evolution has evolved it for that not to work, in effect. It's actively hiding the symptoms of what it's doing, because that's what it's evolved to do, it's evolved to do the same thing no matter whether it's working or not in some sense, or whether each piece of it is working or not.
You do need to get to a different level to see what's going wrong. Mostly the body fixes itself when it's cancering, and so if you can just sort of help it, tilt it in the right direction, probably most cancers would fix themselves if you give them a chance to.
Oddly, though, if you think about that and think about chemotherapy, which is basically the strategy of let's try to poison the cancer, even though we poison you, it seem crazy. Let's try to have a poison that's just enough not to kill you, but to kill the cancer. Well, you're also probably compromising all those mechanisms that fight the cancer, rather than propping them up. In some sense we're probably making the body's job harder in our attempts to treat it.
Mainstream medicine tries to understand what are the mechanisms, what are the pathways, tries to design things that enhance, or block something bad from happening and so on. Proteomics is a new tool that gives you a whole lot more information to do that with, and once you have a whole lot more information, then that lets you do a much more informed dynamic intervention. It helps to give you insight in the actual works of the pathways. That's a very mainstream idea. I don't think anybody would argue with that.
On symbiosis
A really interesting point is that probably a lot of the system that is our healthy body actually doesn't have human DNA at all. It has some other microbial DNA, and we probably are a complicated ecosystem of different types of our own cells, and lots of non-human microbial cells. Once we start looking at the proteome, we'll be looking actually at the conversation of all of those cells, not just the human cells.
Some of those stripes that show up in the picture I showed, we can say, "Oh, that corresponds to this human gene." Some of them, we don't know they might be produced by some combination of other proteins, might be some other organism's protein. Microbial protein would show up in that too, or response to microbes. One of the beauties of this is that we see everything, whether it's of human origin or not. We actually can see it all in the proteome.
On the National Cancer Institute
The leaders of the National Cancer Institute are very keenly aware of how little progress has actually been made in the treatment of cancer. This is something they pay a lot of attention to. They're thinking very laterally in giving funding to people like me to work on cancer. What they've done is to say 'Let's bring in some new kind of thinking to this, and let's have a program where we have physical scientists be the principle investigators, partnered with the co-investigators who are clinicians and biological scientists. I'm partnering with David Agus, for example. But giving money to the physical scientist is a pretty radical idea; you can imagine it is very controversial within the biological community. NCI has started start a few of these centers, and given them five years to work. They need to be interdisciplinary and geographically distributed.
Our center at USC has people all over the United States involved in it, like Cold Spring Harbor, Stanford, Arizona, UT, NYU and Caltech. Typically these labs will have a particular technique for studying biology. It might be measuring proteins within a single cell, or imaging the growth of a tumor. Very often these things are very, very hard to do, and so people have built their career on a new way of sensing proteins inside the cells, and they have teams of graduate students who are refining this technique, and doing variations of this technique, and developing it and so on.
There is about a dozen of these physical sciences centers, each of them is the same kind of structure, there is a physical scientist who is paired with an oncologist, and a group of people that are geographically distributed, applying their techniques to a program of research. In our particular program of research, as I've described, we're trying to take one cancer, study it with all these different techniques, and build a model of how it develops. Different centers have different goals for what they do, but what they all have in common is somebody who is not a biologist trying to use the tools of biology to study cancer in some way.
The proposals for these centers got reviewed by a peer review process, with biologists evaluating at them. One of the reasons that the biologists liked our program was that, surprisingly enough, it's really unusual, even unique for somebody to take one line of cancer and study the same line of cancer with all of these different tools. Typically what people do is study a different cancer with a different tool, and they find the cancer, the type of cells that they can grow in that lab that works well with their tool. The idea of picking a few different specific cell lines, and studying them from a lot of different dimensions, and lots of different scales; I believe that, our proposal probably got funded because the biologists said, "That's going to be useful even though we don't believe this modeling stuff will ever work. Just doing that will be very useful."
A lot of the resources that have been put into cancer have generated the preconditions so that we can do this. I don't feel those resources were wasted in any way. We really understand a huge amount about pathways, and specific mechanisms, so we know a whole lot more about cancer, even though we can't treat it much better in many cases. We have a lot more information, and that information can be made more useful now that we have this proteomic information. This isn't in the category of throw away everything we've done and start over, this is an incremental progress next step that's built on top of an incredible mountain of work.
On how these efforts could fail
There are a couple of ways that I could be wrong, or fail. The timing could be wrong. It could be that we don't have enough of that information built up about the mechanism to interpret these results yet, so it could be that when we really started analyzing we just failed to find the patterns of proteins that correlate with useful things to treat. I would bet then that that's probably just an issue of timing. We're too early, we don't have the right information to bring together with it, or maybe we're not doing the measurement in enough detail.
Maybe it's not these 20 percent changes, maybe it's 2 percent changes, or maybe it's changes in much rarer proteins that we're not measuring by this method, something like that. Maybe it's not in the blood. Maybe most of the information stayed inside the cell. Maybe the action is happening within the cell, not between cells, in which case, probably it will be much longer before it's diagnostically useful in medicine.
Eventually, almost certainly, it will be right. Whether, the timing is right to actually apply this information to medicine in the near future is a risky proposition, and the answer is probably they'll be partially right. There will be some things you can apply it to, and some things that remain mysteries until you look at much more detail of what's going on within each cell, or something like that.
Certainly there are other kinds of molecules that are important, too. It's not just proteins. We know, for example, glucose is important. There are small molecules that are important. Now, most of the small molecules, their production, breakdown, are regulated by proteins, so I'm guessing that if you have all the proteins' state, then you can infer what's going on with the other stuff. But there is state in the body in other things besides proteins. But most of the information seems to be in the proteins.
The body has lots of mechanisms for not only transmitting these proteins, but listening to them. If you look at what these molecular machines do, they talk to each other through these proteins; they say, "If you turn on, produce more of that, or less of that". They're measuring these proteins all the time, and responding to them. That's how your body works, that's how the feedback loops work. Your body is doing proteomics constantly.
It's exciting because for the first time we're really looking at the variables of this complex process, the dynamic variables, which is what life is. That may be too complex for us to comprehend, but at least we're listening in on the conversation. Whether we can understand it or not, we'll start learning in the next few years.
Introduction
Three years ago, Edge collaborated with The Serpentine Gallery in London in a program of "table-top experiments" as part of the Serpentine's Experiment Marathon . This live event was featured along with the Edge/Serpentine collaboration: "What Is Your Formula? Your Equation? Your Algorithm? Formulae For the 21st Century."
Hans Ulrich Obrist, curator of the Serpentine, invited Edge to collaborate in his latest project, The Serpentine Map Marathon, produced in conjunction with DLD (Digital - Life - Design) Saturday and Sunday, 16 – 17 October, at Royal Geographical Society, 1 Kensington Gore, London SW7 2AR (Map). The multi-dimensional Map Marathon features non-stop live presentations by over 50 artists, poets, writers, philosophers, scholars, musicians, architects, designers and scientists. The two-day event takes place in London during Frieze Art Fair week.
In addition to presenting maps by Edge contributors, the Marathon featured a panel of Edge contributors Lewis Wolpert, Armand Leroi, with myself as moderator, at the Royal Geographical Society.
Click on images to enlarge or click here to begin slide show.
 |
 |
 |
 |
 |
 |
 |
 |
Something radically new is in the air: new ways of understanding physical systems, new ways of thinking about thinking that call into question many of our basic assumptions. A realistic biology of the mind, advances in evolutionary biology, physics, information technology, genetics, neurobiology, psychology, engineering, the chemistry of materials: all are questions of critical importance with respect to what it means to be human. For the first time, we have the tools and the will to undertake the scientific study of human nature.
This began in the early seventies, when, as a graduate student at Harvard, evolutionary biologist Robert Trivers wrote five papers that set forth an agenda for a new field: the scientific study of human nature. In the past thirty-five years this work has spawned thousands of scientific experiments, new and important evidence, and exciting new ideas about who and what we are presented in books by scientists such as Richard Dawkins, Daniel C. Dennett, Steven Pinker, and Edward O. Wilson among many others.
In 1975, Wilson, a colleague of Trivers at Harvard, predicted that ethics would someday be taken out of the hands of philosophers and incorporated into the "new synthesis" of evolutionary and biological thinking. He was right.
Scientists engaged in the scientific study of human nature are gaining sway over the scientists and others in disciplines that rely on studying social actions and human cultures independent from their biological foundation.
No where is this more apparent than in the field of moral psychology. Using babies, psychopaths, chimpanzees, fMRI scanners, web surveys, agent-based modeling, and ultimatum games, moral psychology has become a major convergence zone for research in the behavioral sciences.
So what do we have to say? Are we moving toward consensus on some points? What are the most pressing questions for the next five years? And what do we have to offer a world in which so many global and national crises are caused or exacerbated by moral failures and moral conflicts? It seems like everyone is studying morality these days, reaching findings that complement each other more often than they clash.
Culture is humankind’s biological strategy, according to Roy F. Baumeister, and so human nature was shaped by an evolutionary process that selected in favor of traits conducive to this new, advanced kind of social life (culture). To him, therefore, studies of brain processes will augment rather than replace other approaches to studying human behavior, and he fears that the widespread neglect of the interpersonal dimension will compromise our understanding of human nature. Morality is ultimately a system of rules that enables groups of people to live together in reasonable harmony. Among other things, culture seeks to replace aggression with morals and laws as the primary means to solve the conflicts that inevitably arise in social life. Baumeister’s work has explored such morally relevant topics as evil, self-control, choice, and free will. [More]
According to Yale psychologist Paul Bloom, humans are born with a hard-wired morality. A deep sense of good and evil is bred in the bone. His research shows that babies and toddlers can judge the goodness and badness of others' actions; they want to reward the good and punish the bad; they act to help those in distress; they feel guilt, shame, pride, and righteous anger. [More]
Harvard cognitive neuroscientist and philosopher Joshua D. Greene sees our biggest social problems — war, terrorism, the destruction of the environment, etc. — arising from our unwitting tendency to apply paleolithic moral thinking (also known as "common sense") to the complex problems of modern life. Our brains trick us into thinking that we have Moral Truth on our side when in fact we don't, and blind us to important truths that our brains were not designed to appreciate. [More]
University of Virginia psychologist Jonathan Haidt's research indicates that morality is a social construction which has evolved out of raw materials provided by five (or more) innate "psychological" foundations: Harm, Fairness, Ingroup, Authority, and Purity. Highly educated liberals generally rely upon and endorse only the first two foundations, whereas people who are more conservative, more religious, or of lower social class usually rely upon and endorse all five foundations. [More]
The failure of science to address questions of meaning, morality, and values, notes neuroscientist Sam Harris, has become the primary justification for religious faith. In doubting our ability to address questions of meaning and morality through rational argument and scientific inquiry, we offer a mandate to religious dogmatism, superstition, and sectarian conflict. The greater the doubt, the greater the impetus to nurture divisive delusions. [More]
A lot of Yale experimental philosopher Joshua Knobe's recent research has been concerned with the impact of people's moral judgments on their intuitions about questions that might initially appear to be entirely independent of morality (questions about intention, causation, etc.). It has often been suggested that people's basic approach to thinking about such questions is best understood as being something like a scientific theory. He has offered a somewhat different view, according to which people's ordinary way of understanding the world is actually infused through and through with moral considerations. He is arguably most widely known for what has come to be called "the Knobe effect" or the "Side-Effect Effect." [More]
NYU psychologist Elizabeth Phelps investigates the brain activity underlying memory and emotion. Much of Phelps' research has focused on the phenomenon of "learned fear," a tendency of animals to fear situations associated with frightening events. Her primary focus has been to understand how human learning and memory are changed by emotion and to investigate the neural systems mediating their interactions. A recent study published in Nature by Phelps and her colleagues, shows how fearful memories can be wiped out for at least a year using a drug-free technique that exploits the way that human brains store and recall memories. [More]
Disgust has been keeping Cornell psychologist David Pizarro particularly busy, as it has been implicated by many as an emotion that plays a large role in many moral judgments. His lab results have shown that an increased tendency to experience disgust (as measured using the Disgust Sensitivity Scale, developed by Jon Haidt and colleagues), is related to political orientation. [More]
Each of the above participants led a 45-minute session on Day One that consisted of a 25-minute talk. Day Two consisted of two 90-minute open discussions on "The Science of Morality", intended as a starting point to begin work on a consensus document on the state of moral psychology to be published onEdge in the near future.
Among the members of the press in attendance were: Sharon Begley, Newsweek, Drake Bennett, Ideas, Boston Globe, David Brooks, OpEd Columnist, New York Times, Daniel Engber, Slate, Amanda Gefter, Opinion Editor,New Scientist, Jordan Mejias, Frankfurter Allgemeine Zeitung,Gary Stix, Scientific American, Pamela Weintraub, Discover Magazine.
The New Science of Morality, Part 1

[JONATHAN HAIDT:] As the first speaker, I'd like to thank the Edge Foundation for bringing us all together, and bringing us all together in this beautiful place. I'm looking forward to having these conversations with all of you.
I was recently at a conference on moral development, and a prominent Kohlbergian moral psychologist stood up and said, "Moral psychology is dying." And I thought, well, maybe in your neighborhood property values are plummeting, but in the rest of the city, we are going through a renaissance. We are in a golden age.
The New Science of Morality, Part 2

[JOSHUA D. GREENE:] Now, it's true that, as scientists, our basic job is to describe the world as it is. But I don't think that that's the only thing that matters. In fact, I think the reason why we're here, the reason why we think this is such an exciting topic, is not that we think that the new moral psychology is going to cure cancer. Rather, we think that understanding this aspect of human nature is going to perhaps change the way we think and change the way we respond to important problems and issues in the real world. If all we were going to do is just describe how people think and never do anything with it, never use our knowledge to change the way we relate to our problems, then I don't think there would be much of a payoff. I think that applying our scientific knowledge to real problems is the payoff.
The New Science of Morality, Part 3

[SAM HARRIS:] ...I think we should differentiate three projects that seem to me to be easily conflated, but which are distinct and independently worthy endeavors. The first project is to understand what people do in the name of "morality." We can look at the world, witnessing all of the diverse behaviors, rules, cultural artifacts, and morally salient emotions like empathy and disgust, and we can study how these things play out in human communities, both in our time and throughout history. We can examine all these phenomena in as nonjudgmental a way as possible and seek to understand them. We can understand them in evolutionary terms, and we can understand them in psychological and neurobiological terms, as they arise in the present. And we can call the resulting data and the entire effort a "science of morality". This would be a purely descriptive science of the sort that I hear Jonathan Haidt advocating.
The New Science of Morality, Part 4

[ROY BAUMEISTER:] And so that said, in terms of trying to understand human nature, well, and morality too, nature and culture certainly combine in some ways to do this, and I'd put these together in a slightly different way, it's not nature's over here and culture's over there and they're both pulling us in different directions. Rather, nature made us for culture. I'm convinced that the distinctively human aspects of psychology, the human aspects of evolution were adaptations to enable us to have this new and better kind of social life, namely culture.
Culture is our biological strategy. It's a new and better way of relating to each other, based on shared information and division of labor, interlocking roles and things like that. And it's worked. It's how we solve the problems of survival and reproduction, and it's worked pretty well for us in that regard. And so the distinctively human traits are ones often there to make this new kind of social life work.
Now, where does this leave us with morality?
The New Science of Morality, Part 5

[PAUL BLOOM:] What I want to do today is talk about some ideas I've been exploring concerning the origin of human kindness. And I'll begin with a story that Sarah Hrdy tells at the beginning of her excellent new book, "Mothers And Others." She describes herself flying on an airplane. It’s a crowded airplane, and she's flying coach. She's waits in line to get to her seat; later in the flight, food is going around, but she's not the first person to be served; other people are getting their meals ahead of her. And there's a crying baby. The mother's soothing the baby, the person next to them is trying to hide his annoyance, other people are coo-cooing the baby, and so on.
As Hrdy points out, this is entirely unexceptional. Billions of people fly each year, and this is how most flights are. But she then imagines what would happen if every individual on the plane was transformed into a chimp. Chaos would reign. By the time the plane landed, there'd be body parts all over the aisles, and the baby would be lucky to make it out alive.
The point here is that people are nicer than chimps.
The New Science of Morality, Part 6

[DAVID PIZARRO:] What I want to talk about is piggybacking off of the end of Paul's talk, where he started to speak a little bit about the debate that we've had in moral psychology and in philosophy, on the role of reason and emotion in moral judgment. I'm going to keep my claim simple, but I want to argue against a view that probably nobody here has, (because we're all very sophisticated), but it's often spoken of emotion and reason as being at odds with each other — in a sense that to the extent that emotion is active, reason is not active, and to the extent that reason is active, emotion is not active. (By emotion here, I mean, broadly speaking, affective influences).
I think that this view is mistaken (although it is certainly the case sometimes). The interaction between these two is much more interesting. So I'm going to talk a bit about some studies that we've done. Some of them have been published, and a couple of them haven't (because they're probably too inappropriate to publish anywhere, but not too inappropriate to speak to this audience). They are on the role of emotive forces in shaping our moral judgment. I use the term "emotive," because they are about motivation and how motivation affects the reasoning process when it comes to moral judgment.
The New Science of Morality, Part 7

[ELIZABETH PHELPS:] In spite of these beliefs I do think about decisions as reasoned or instinctual when I'm thinking about them for myself. And this has obviously been a very powerful way of thinking about how we do things because it goes back to earliest written thoughts. We have reason, we have emotion, and these two things can compete. And some are unique to humans and others are shared with other species.
And economists, when thinking about decisions, have also adopted what we call a dual system approach. This is obviously a different dual system approach and here I'm focusing mostly on Kahneman's System 1 and System 2. As probably everybody in this room knows Kahneman and Tversky showed that there were a number of ways in which we make decisions that didn't seem to be completely consistent with classical economic theory and easy to explain. And they proposed Prospect Theory and suggested that we actually have two systems we use when making decisions, one of which we call reason, one of which we call intuition.
Kahneman didn't say emotion. He didn't equate emotion with intuition.
The New Science of Morality, Part 8

[JOSHUA KNOBE:] ...what's really exciting about this new work is not so much just the very idea of philosophers doing experiments but rather the particular things that these people ended up showing. When these people went out and started doing these experimental studies, they didn't end up finding results that conformed to the traditional picture. They didn't find that there was a kind of initial stage in which people just figured out, on a factual level, what was going on in a situation, followed by a subsequent stage in which they used that information in order to make a moral judgment. Rather they really seemed to be finding exactly the opposite.
What they seemed to be finding is that people's moral judgments were influencing the process from the very beginning, so that people's whole way of making sense of their world seemed to be suffused through and through with moral considerations. In this sense, our ordinary way of making sense of the world really seems to look very, very deeply different from a kind of scientific perspective on the world. It seems to be value-laden in this really fundamental sense.
EDGE IN THE NEWS
BOSTON GLOBE
August 15, 2010
IDEAS
The surprising moral force of disgust
By Drake Bennett
...Psychologists like Haidt are leading a wave of research into the so-called moral emotions — not just disgust, but others like anger and compassion — and the role those feelings play in how we form moral codes and apply them in our daily lives. A few, like Haidt, go so far as to claim that all the world's moral systems can best be characterized not by what their adherents believe, but what emotions they rely on.
There is deep skepticism in parts of the psychology world about claims like these. And even within the movement there is a lively debate over how much power moral reasoning has — whether our behavior is driven by thinking and reasoning, or whether thinking and reasoning are nothing more than ornate rationalizations of what our emotions ineluctably drive us to do. Some argue that morality is simply how human beings and societies explain the peculiar tendencies and biases that evolved to help our ancestors survive in a world very different from ours.
A few of the leading researchers in the new field met late last month at a small conference in western Connecticut, hosted by the Edge Foundation, to present their work and discuss the implications. Among the points they debated was whether their work should be seen as merely descriptive, or whether it should also be a tool for evaluating religions and moral systems and deciding which were more and less legitimate — an idea that would be deeply offensive to religious believers around the world.
But even doing the research in the first place is a radical step. The agnosticism central to scientific inquiry is part of what feels so dangerous to philosophers and theologians. By telling a story in which morality grows out of the vagaries of human evolution, the new moral psychologists threaten the claim of universality on which most moral systems depend — the idea that certain things are simply right, others simply wrong. If the evolutionary story about the moral emotions is correct, then human beings, by being a less social species or even having a significantly different prehistoric diet, might have ended up today with an entirely different set of religions and ethical codes. Or we might never have evolved the concept of morals at all. ...
THE ATLANTIC
July 29, 2010
Alexis Madrigal
University of Virginia moral psychologist Jonathan Haidt delivered an absolutely dynamite talk on new advances in his field last week. The video and a transcript have been posted by Edge.org, a loose consortium of very smart people run by John Brockman. Haidt whips us through centuries of moral thought, recent evolutionary psychology, and discloses which two papers every single psychology student should have to read. Through it all, he's funny, erudite, and understandable. Here, we excerpt a few paragraphs from his conclusion, in which Haidt tells us how to think about our moral minds: ...
FRANKFURTER ALLGEMEINE ZEITUNG
July 28, 2010
FEUILLETON
Moral reasoning
SOLEMN HIGH MASS IN THE TEMPLE OF REASON
How do you train a moral muscle? American researchers take their first steps on the path to a science of morality without God hypothesis. The last word should have the reason.
By Jordan Mejias
[Google translation:]
28th July 2010 One was missing and had he turned up, the illustrious company would have had nothing more to discuss and think. Even John Brockman, literary agent, and guru of the third culture, it could not move, stop by in his salon, which he every summer from the virtuality of the Internet, click on edge.org moved, in a New England idyl. There, in the green countryside of Washington, Connecticut, it was time to morality as a new science. When new it was announced, because their devoted not philosophers and theologians, but psychologists, biologists, neurologists, and at most such philosophers, based on experiments and the insights of brain research. They all had to admit, even to be on the search, but they missed not one who lacked the authority in matters of morality: God.
The secular science dominated the conference. As it should come to an end, however, a consensus first, were the conclusions apart properly. Even on the question of whether religion should be regarded as part of evolution, remained out of the clear answer. Agreement, the participants were at least that is to renounce God. Him, the unanimous result of her certainly has not been completed or not may be locked investigations, did not owe the man morality. That it is innate in him, but did so categorically not allege any. Only on the findings that morality is a natural phenomenon, there was agreement, even if only to a certain degree. For, should be understood not only the surely. Besides nature makes itself in morality and the culture just noticeable, and where the effect of one ends and the other begins, is anything but settled.
Better be nice
In a baby science, as Elizabeth Phelps, a neuroscientist at New York University, called the moral psychology may by way of surprise not much groping. How about some with free will, will still remain for the foreseeable future a mystery. Moral instincts was, after all, with some certainty Roy Baumeister, a social psychologist at Florida State University, are not built into us. We are only given the ability to acquire systems of morality. Gives us to be altruistic, we are selfish by nature, benefits. It is moral to be compared with a muscle, the fatigue, but can also be strengthened through regular training. What sounds easier than is done, if not clear what is to train as well. A moral center that we can selectively edit points, our brain does not occur.
But amazingly, with all that we are nice to each other are forced reproduction, and Paul Bloom, a psychologist at Yale, is noticed. Obviously, we have realized that our lives more comfortable when others do not fight us. Factors of Nettigkeitswachstums Bloom also recognizes in capitalism that will work better with nice people, and world religions, which act in large groups and their dynamics as it used to strangers to meet each other favorably. The fact that we have developed over the millennia morally beneficial, holds not only he has been proved. Even the neurologist Sam Harris, author of "The Moral Landscape. How Science Can Determine Human Values "(Free Press), wants to make this progress not immoral monsters like Hitler and Stalin spoil. ...
[...Continue: German language original | Google translation]
ANDREW SULLIVAN — THE DAILY DISH
25 JUL 2010
Edge held a seminar on morality. Here's Joshua Knobe:
Over the past few years, a series of recent experimental studies have reexamined the ways in which people answer seemingly ordinary questions about human behavior. Did this person act intentionally? What did her actions cause? Did she make people happy or unhappy? It had long been assumed that people's answers to these questions somehow preceded all moral thinking, but the latest research has been moving in a radically different direction. It is beginning to appear that people's whole way of making sense of the world might be suffused with moral judgment, so that people's moral beliefs can actually transform their most basic understanding of what is happening in a situation.
David Brooks' illuminating column on this topic covered the same ground:
...
...Advantage Locke over Hobbes.
THE NEW YORK TIMES
July 23, 2010
OP-ED COLUMNIST
Scientific research is showing that we are born with an innate moral sense.
By DAVID BROOKS
Washington, Conn.
Where does our sense of right and wrong come from? Most people think it is a gift from God, who revealed His laws and elevates us with His love. A smaller number think that we figure the rules out for ourselves, using our capacity to reason and choosing a philosophical system to live by.
Moral naturalists, on the other hand, believe that we have moral sentiments that have merged from a long history of relationships. To learn about morality, you don't rely upon revelation or metaphysics; you observe people as they live.
This week a group of moral naturalists gathered in Connecticut at a conference organized by the Edge Foundation. ...
By the time humans came around, evolution had forged a pretty firm foundation for a moral sense. Jonathan Haidt of the University of Virginia argues that this moral sense is like our sense of taste. We have natural receptors that help us pick up sweetness and saltiness. In the same way, we have natural receptors that help us recognize fairness and cruelty. Just as a few universal tastes can grow into many different cuisines, a few moral senses can grow into many different moral cultures.
Paul Bloom of Yale noted that this moral sense can be observed early in life. Bloom and his colleagues conducted an experiment in which they showed babies a scene featuring one figure struggling to climb a hill, another figure trying to help it, and a third trying to hinder it. ...
THE REALITY CLUB
QUESTIONS FOR "THE MORAL NINE" FROM THE EDGE COMMUNITY
Howard Gardner, Geoffrey Miller, Brian Eno, James Fowler, Rebecca Mackinnon, Jaron Lanier, Eva Wisten, Brian Knutson, Andrian Kreye, Anonymous, Alison Gopnik, Robert Trivers, Randoph Nesse, M.D.
HOWARD GARDNER
Psychologist, Harvard University; Author, Changing Minds
Enlightenment ideas were the product of white male Christians living in the 18th century. They form the basis of the Universal Declaration of Human Rights and other Western-inflected documents. But in our global world, Confucian societies and Islamic societies have their own guidelines about progress, individuality, democratic processes, human obligations. In numbers they represent more of humanity and are likely to become even more numerous in this century. What do the human sciences have to contribute to an understanding of these 'multiple voices' ? Can they combined harmoniously or are there unbridgeable gaps?
GEOFFREY MILLER
Evolutionary Psychologist, University of New Mexico; Author, Spent: Sex, Evolution, and Consumer Behavior
1) Many people become vegans, protect animal rights, and care about the long-term future of the environment. It seems hard to explain these 'green virtues' in terms of the usual evolutionary-psychology selection pressures: reciprocity, kin selection, group selection -- so how can we explain their popularity (or unpopularity?)
2) What are the main sex differences in human morality, and why?
3) What role did costly signaling play in the evolution of human morality (i.e. 'showing off' certain moral virtues' to attract mates, friends, or allies, or to intimidate rival individuals or competing groups)?
4) Given the utility of 'adaptive self-deception' in human evolution -- one part of the mind not knowing what adaptive strategies another part is pursuing -- what could it mean to have the moral virtue of 'integrity' for an evolved being?
5) Why do all 'mental illnesses' (depression, mania, schizophrenia, borderline, psychopathy, narcissism, mental retardation, etc.) reduce altruism, compassion, and loving-kindness? Is this partly why they are recognized as mental illnesses?
BRIAN ENO
Artist; Composer; Recording Producer: U2, Cold Play, Talking Heads, Paul Simon; Recording Artist
Is morality a human invention - a way of trying to stabilise human societies and make them coherent - or is there evidence of a more fundamental sense of morality in creatures other than humans?
Another way of asking this question is: are there moral concepts that are not specifically human?
Yet another way of asking this is: are moral concepts specifically the province of human brains? And, if they are, is there any basis for suggesting that there are any 'absolute' moral precepts?
Or: do any other creatures exhibit signs of 'honour' or 'shame'??
JAMES FOWLER
Political Scientist, University of California, San Diego; Coauthor, Connected
Given recent evidence about the power of social networks, what is our personal responsibility to our friends' friends?
REBECCA MACKINNON
Blogger & Cofounder, Global Voices Online; Former CNN journalist and head of CNN bureaus in Beijing & Tokyo; Visiting Fellow, Princeton University's Center for Information Technology Policy
Does the human race require a major moral evolution in order to survive? Isn't part of the problem that our intelligence has vastly out-evolved our morality, which is still stuck back in the paleolithic age? Is there anything we can do? Or is this the tragic flaw that dooms us? Might technology help to facilitate or speed up our moral evolution, as some say technology is already doing for human intelligence? We have artificial intelligence and augmented reality. What about artificial or augmented morality?
JARON LANIER
Musician, Computer Scientist; Pioneer of Virtural Reality; Author, You Are Not A Gadget: A Manifesto
A crucial topic is how group interactions change moral perception. To what degree are there clan-oriented processes inherent in the human brain? In particular, how can well-informed software designs for network-mediated social experience play a role in changing behavior and values? Is there anything specific that can be done to reduce mob-like phenomena, as is spawned in online forums like 4chan's /b/, without resorting to degrees of imposed control? This is where a science of moral psychology could inform engineering.
EVA WISTEN
Journalist; Author, Single in Manhattan
What's would be a good definition - a few examples - of common moral sense? How does an averagely moral human think and behave (it's easy to paint a picture of the actions of an immoral person...) Now, how can this be expanded?
Could an understanding/acceptance of the idea that we are all having unconscious instincts for what's right and wrong replace the idea of religion as necessary for moral behavior?
What tends to be the hierarchy of "blinders" - the arguments we, consciously or unconsciously, use to relabel exploitative acts as good? (I did it for God, I did it for the German People, I did it for Jodie Foster...) What evolutionary purpose have they filled?
BRIAN KNUTSON
Psychologist & Neuroscientist, Stanford
What is the difference between morality and emotion? How can scientists distinguish between the two (or should they)? Why has Western culture been so historically reluctant to recognize emotion as a major influence on moral judgments?
ANDRIAN KREYE?
Feuilleton Editor, Sueddutsche Zeitung
Is there a fine line or a wide gap between morality and ideology?
ANONYMOUS
1. Some of the new literature on moral psychology feels like traditional discussions of ethics with a few numbers attached from surveys; almost like old ideas in a new can. As an outsider I'd be curious to know what's really new here. Specifically, if William James were resurrected what might be the new findings we could explain to him that would astound him or fundamentally change his way of thinking?
2. Is there a reason to believe there is such a thing as moral psychology that transcends upbringing and culture? Are we really studying a fundamental feature of the mind or simply the outcome of a social process?
ALISON GOPNIK
Psychologist, UC, Berkeley; Author, The Philosophical Baby
Many people have proposed an evolutionary psychology/ nativist view of moral capacities. But surely one of the most dramatic and obvious features of our moral capacities is their capacity for change and even radical transformation with new experiences. At the same time this transformation isn't just random but seems to have a progressive quality. Its analogous to science which presents similar challenges to a nativist view. And even young children are empirically, capable of this kind of change in both domains. How do we get to new and better conceptions of the world, cognitive or moral, if the nativists are right?
ROBERT TRIVERS
Evolutionary Biologist, Rutgers University; Coauthor, Genes In Conflict: The Biology of Selfish Genetic Elements
Shame.
What is it? When does it occur? What function does it serve? How is it related, if at all, to guilt? Is it related to "morality" and if so how?
Key point, John, is that shame is a complex mixture of self and other: Tiger Woods SHAMES his wife in public — he may likewise be ashamed.
If i fuck a goat i may feel ashamed if someone saw it, but absent harm to the goat, not clear how i should respond if i alone witness it.
A NEW AGE OF WONDER
In a 2009 talk at the Bristol Festival of Ideas, Freeman Dyson articulated a vision for the future. Referencing The Age Of Wonder, by Richard Holmes, which was centered on chemistry and poetry, Dyson pointed out that we are entering a new Age of Wonder, which dominated by computational biology.
"A new generation of artists," Dyson said, "writing genomes as fluently as Blake and Byron wrote verses, might create an abundance of new flowers and fruit and trees and birds to enrich the ecology of our planet. Most of these artists would be amateurs, but they would be in close touch with science, like the poets of the earlier Age of Wonder. The new Age of Wonder might bring together wealthy entrepreneurs ... and a worldwide community of gardeners and farmers and breeders, working together to make the planet beautiful as well as fertile, hospitable to hummingbirds as well as to humans."
The leaders of the new Age of Wonder, Dyson noted, include "biology wizards" Kary Mullis, Craig Venter, medical engineer Dean Kamen; and "computer wizards" Larry Page, Sergey Brin, and Charles Simonyi, and John Brockman and Katinka Matson, the cofounders of Edge, the nexus of this intellectual activity.
A number of Edge contributors were among the speakers at the recent TED conference (TED 2010) in Long Beach: Stewart Brand,Nicholas Christakis, George Church, Denis Dutton, Sam Harris, Daniel Kahneman, Benoit Mandelbrot, Nathan Myhrvold, Michael Shermer, and even one of the first speakers at the Reality Club of the early 80s, Stephen Wolfram. And many more individuals in the Edge community as well as friends were in attendance. Come, take a walk with me.
Below is a link to Daniel Kahneman's recently posted TED Talk — featuring the superb video production values that are becoming the hallmark of the TED Talks — along with links to Kahneman's relevant Edge contributions.
—JB