Edge Master Class 2010: W. DANIEL HILLIS ON "CANCERING" [1]
We make a mistake when we think of cancer as a noun. It is not something you have, it is something you do. Your body is probably cancering all the time. What keeps it under control is a conversation that is happening between your cells, and the language of that conversation is proteins. Proteomics will allow us to listen in on that conversation, and that will lead to much better way to treat cancer.

Introduction
by John Brockman
The leaders of the National Cancer Institute," says Danny Hillis, "are very keenly aware of how little progress has actually been made in the treatment of cancer. This is something they pay a lot of attention to. They're thinking very laterally in giving funding to people like me to work on cancer."
"What they've said is 'Let's bring some new kinds of thinking to this, and create a program where we have physical scientists be the principle investigators, partnered with the co-investigators who are clinicians and biological scientists.' I'm partnering with David Agus, for example. But giving money to the physical scientist is a pretty radical idea, you can imagine it is very controversial within the biological community."
"NCI has started a few of these centers, and given them five years to work. They need to be interdisciplinary and geographically distributed. Our center at USC has people all over the United States involved in it, like Cold Spring Harbor, Stanford, Arizona, UT, NYU and CalTech.
"As a result, Hillis is the newly appointed professor of research medicine at the Keck School of Medicine at the University of Southern California (USC). And he is the principal investigator of a five-year government program on cancer.
The University of Southern California Physical Sciences-Oncology Center's (USC PS-OC) overall goal is to thoroughly understand therapeutic response.
Investigators will establish a predictive model of cancer that they can utilize to determine tumor steady state growth and drug response, particularly those involved in the hematological malignancies of acute myeloid leukemia and non-Hodgkin lymphoma. Furthermore, multi-scale physical measurements will be unified with sophisticated modeling approaches to facilitate the development of a model that can derive the tumor's traits during its growth and after any distress, such as chemotherapeutic treatment. These investigators will apply pioneering measurement platforms to resolve real-time protein interactions and protein abundance and to characterize protein modifications. Appropriately, these studies will also address tumor and host response to therapy using a systems approach. Overall, the predictive tumor response model should enable clinicians to determine the most efficacious therapies a priori and reduce deleterious side effects.
Hillis continues..."We misunderstand cancer by making it a noun. Instead of saying, "My house has water", we say, "My plumbing is leaking." Instead of saying, "I have cancer", we should say, "I am cancering." The truth of the matter is we're probably cancering all the time, and our body is checking it in various ways, so we're not cancering out of control. Probably every house has a few leaky faucets, but it doesn't matter much because there are processes that are mitigating that by draining the leaks. Cancer is probably something like that.
"In order to understand what's actually going on, we have to look at the level of the things that are actually happening, and that level is proteomics. Now that we can actually measure that conversation between the parts, we're going to start building up a model that's a cause-and-effect model: This signal causes this to happen, that causes that to happen. Maybe we will not understand to the level of the molecular mechanism but we can have a kind of cause-and-effect picture of the process. More like we do in sociology or economics.
"Last year's EdgeMaster Class in Los Angeles featured George Church and Craig Venter lecturing on Synthetic Genomics. Hillis points out that the genome is used to construct things, and that it's not the best place for analysis of what's going on. "Certainly," he says, "there are times it is useful, but I don't think that's where most of the information is.
If you think in terms of computer models, think of proteomics as a debugging tool for genomics programs. "When you write a computer program, the first thing you do is you try to run it, and it almost always has a bug in it, so you see what happens, and you debug it, you stop it in the middle of running, and you see what the state of the system is, and you understand what your bug is, and then you change the program. The proteome is the state".
"Proteomics was made possible by genomics. It builds on top of genomics. I guess it's true in some theoretical sense that eventually you might not even bother to look at the genome if you can see the whole proteome, but in practice, it's been very important."
"The genome is the instructions for the cell. That's very important if you want to do manipulation. If you want to actually affect the pathway, then that is the level at which you need to manipulate things. You want to knock out a gene, or modify a gene. Experimentally, being able to read and write the genome is incredibly important. But if you want to use it as a diagnostic for what's going wrong with a particular individual, it will be unusual for that information to be in the genome."

I am talking to Hillis in Villa Miravelle, an historic, exotic mansion with an interesting history, built in 1884 in Saint Helena, CA, by Mexican-American Tiburcio Parrott. In 1974 Spring Mountain Vineyard and winery were established on the surrounding property.

Swiss financier Jacob E. "Jacqui" Safra [2] acquired the Spring Mountain Vineyard in 1991 and through acquisitions expanded the property to 225 acres of vineyard on an 850 acre estate, now the largest vineyard in terms of contiguous acreage in the Napa Valley. Safra, a member of the Edge community, graciously offered the use of Spring Mountain Vineyard [3] as the venue for the Edge Master Class 2010.
This year, to try something different, Edge ran a conference on the East Coast in July (See "The New Science of Morality [4]") There was no intent to run the usual Master Class in California, until Safra came forth with his generous offer, followed by a conversation I had with Danny Hillis a week ago on "proteomics". Given his excitement about the prospects of his new research program coupled by his track record as the man who broke the von Neumann bottleneck to give us the massively parallel computer, I didn't hesitate to announce this event.
The event occurred with one week's notice, the same week as the "Sci-FOO Camp" at Google and the first "Techonomy" conference at Lake Tahoe, both interesting, even exciting events. In the end, among all the usual suspects, it was Hillis, Stewart Brand [5], and myself who showed up for a weekend at the most exquisite vineyard property in California (See photos [6] below). The weather was beautiful. We were surrounded by dozens of bottles of Safra's prize-winning Spring Mountain Elivette (2005).

Stewart Brand [5], John Brockman [7], Daniel Hillis [8]
We a 2-part Edge Master Class which is available below in three formats: streaming viideo (two one-hour talks), audio download, and text (including a printable text file of Parts I & II) .
- JB
W. DANIEL ("DANNY") HILLIS is Chairman and Chief Technology Officer of Applied Minds, a research and development company creating a range of new products and services in software, entertainment, electronics, biotechnology and mechanical design. Hillis is also Judge Widney Professor of Engineering and Medicine of the University of Southern California (USC), professor of research medicine at the Keck School of Medicine, and research professor of engineering at the Viterbi School of Engineering. Previously, Hillis was Vice President, Research and Development at Walt Disney Imagineering, and Disney Fellow. He developed new technologies and business strategies for Disney's theme parks, television, motion pictures, Internet and consumer products businesses.
An inventor, scientist, engineer, author, and visionary, Hillis pioneered the concept of parallel computers that is now the basis for most supercomputers, as well as the RAID disk array technology used to store large databases. He holds over 150 U.S. patents, covering parallel computers, disk arrays, forgery prevention methods, and various electronic and mechanical devices.
As a student at MIT, Hillis began to study the physical limitations of computation and the possibility of building highly parallel computers. This work culminated in 1985 with the design of a massively parallel computer with 64,000 processors. He named it the Connection Machine. During this period at MIT Hillis co-founded Thinking Machines Corp. to produce and market the Connection Machine.
Thinking Machines Corp. was the leading innovator in massive parallel supercomputers and RAID disk arrays. In addition to designing the company's major products, Hillis worked closely with his customers in applying parallel computers to problems in astrophysics, aircraft design, financial analysis, genetics, computer graphics, medical imaging, image understanding, neurobiology, materials science, cryptography and subatomic physics. At Thinking Machines, he built a technical team comprised of scientists and engineers that were widely acknowledged to have been among the best in the industry.
In 2005, Hillis and others from Applied Minds initiated Metaweb Technologies to develop a semantic data storage infrastructure for the Internet, and Freebase, an "open, shared database of the world's knowledge". That company was recently acquired by Google.
Hillis has published scientific papers in journals such as Science, Nature, Modern Biology, Communications of the ACM andInternational Journal of Theoretical Physics and is an editor of several other scientific journals, including Artificial Life, Complexity, Complex Systems, Future Generation Computer Systems andApplied Mathematics. He has also written extensively on technology and its implications for publications such as Newsweek, Wired, Forbes ASAP and Scientific American. He is the author of The Pattern on the Stone. He is a Member of the National Academy of Engineering, a Fellow of the Association of Computing Machinery, a Fellow of the International Leadership Forum, and a Fellow of the American Academy of Arts and Sciences. He is Co-Chair of Long Now Foundation and the designer of a 10,000-year mechanical clock.
Danny Hillis's Edge Bio Page [9]
We are pleased to present below the entire 2-part Edge Master Class which is available below in three formats: streaming viideo, audio download, and text.
![]()
CANCERING
Listening In On The Body's Proteomic Conversation (Part I)
Right now, I am asking a lot of questions about cancer, but I probably should explain how I got to that point, why somebody who's mostly interested in complexity, and computers, and designing machines, and engineering, should be interested in cancer. I'll tell you a little bit about cancer, but before I tell you about that, I'm going to tell you about proteomics, and before I tell you about proteomics, I want to get you to think about genomics differently because people have heard a lot about genes, and genomics in the last few years, and it's probably given them a misleading idea about what's important, how diseases work, and so on. ...
~~~
Text (Part I) [10]
Printable text file (Parts I & II) [11]
You've probably heard the genome described as like a blueprint for producing an organism. That's a very misleading analogy because a blueprint is interesting because it says how everything is connected, and how the parts relate to each other. In fact, the genome, at least the part of the genome that we understand how to read, actually doesn't tell you that at all. It's kind of a list of the parts. It does have some control information on it about when different parts should be made, but for the most part we don't know how to read that control information right now. What we know how to read is the parts list. While that's a very useful thing, it's probably not the most important thing that we need to know to understand what's going on.CANCERING: Listening In On The Body's Proteomic Conversation (PART I)
W. Daniel Hillis
Right now, I am asking a lot of questions about cancer, but I probably should explain how I got to that point, why somebody who's mostly interested in complexity, and computers, and designing machines, and engineering, should be interested in cancer. I'll tell you a little bit about what I am doing in cancer, but before I tell you about that, I'm going to tell you about proteomics. Before I tell you about proteomics, I want to get you to think about genomics differently because people have heard a lot about genes and genomics in the last few years, and it's probably given them a misleading idea about what's important, how diseases work, and so on.
Let me start by talking about genes, and giving you a different way of looking at genes, I want to start by clearing up, well maybe not misunderstandings, but putting a different emphasis on how genes work. That will explain why I'm interested in proteomics, and that will explain why I'm interested in cancer.
An analogy might be restaurants. Let's say you were trying to understand the difference between a great restaurant and a bad restaurant, and what you had to work with was a list of the ingredients that they had in their storehouse. Sure enough, if you snuck in at night and looked at their inventory list, you might be able to tell some things about the restaurant. You could probably tell the difference between a French restaurant, and a Chinese restaurant just by the ingredients list. And indeed, you can tell the difference between a European person, and an Oriental person just by looking at their ingredients list, but you probably can't tell a lot about what their personality is like.
Now, sometimes you can tell about defects, so if a restaurant was completely missing salt, or they only had lard for oil, you could say, "Well, this restaurant might be improved if they started using salt, or if they had some butter instead of lard", or something like that. So there might be some gross things that you can tell about inadequacies, that they were missing a key ingredient, something was broken about the ingredients list, but to really understand whether the food was good or not, or how they were making food, you really have to watch what's going on in the kitchen, and watch the process. You have to actually watch the dynamic process; the list of ingredients doesn't work.
The way I think about this is more like computer programs. The genome is like a listing of your operating system, but missing all the control information, so missing all the jumps, and things like that, and that's a fairly useless thing to have if you're trying to figure out, if you're trying to debug a program. It's not totally useless, but it's not that informative. What a programmer would really want to know is they'd like to dynamically look at what's going on inside the machine, what's getting loaded into the registers. That kind of dynamic trace is the much more useful thing for debugging them in kind of a partial listing of the code. If I put it that way, you might ask what's the big deal about the genome, why all the excitement about genomics?
I think it has a couple of historical reasons. One of them is the gene is the great theoretical triumph of biology, it's the one kind of theoretical construct that was predicted. Like the physicists predicted there ought to be a positron, and when they looked, there was a positron. That happens all the time in physics. The equation says there should be a black hole, and we look, and we find black holes. In biology, that almost never happens, and the great dramatic example of it happening was genes. So genes were kind of theoretically predicted by Mendel and they were the core of what Darwin needed, that unit of inheritance. Then Watson and Crick looked and actually discovered it! It was like actually finding the black hole that was predicted. That was in some sense the most exciting thing that ever happened in biology, and since it so stands out, there's nothing close to that, it almost has a religious significance in biology. It is the triumph of the one great theory.
The other thing about it, a practical thing, is that it turned out that people like Kary Mullis worked out this very neat way with tools that the biologists had on their bench, so they could actually measure a gene. In fact, you can almost do genetics in your own kitchen with a few extra pieces of equipment, if you have the right enzymes around. You heat something up, and cool it down, and heat it up, and cool it down, and then you pour it in some jello, run an electric field across it and you actually get a read-out of this nice, digital picture.
So not only was it a theoretical construct that had been predicted by biology, but it was also accessible to experiments with the stuff that people had lying around in their labs. Of course, now we sequence genes with much more sophisticated equipment that does it much more rapidly. But it got its start because everybody could do it in their lab. They could see the genes, so everybody could get in the genetics business immediately, and start getting really interesting genetics results.
For instance, the field of zoology was transformed by being able to tell what's related to what, like kind of the trick of telling the difference between the French restaurant and the Chinese restaurant. By looking at the ingredients you could find the complete tree of family relationships, and so there's a huge amount of good science that suddenly became possible. You could get a lot of hard data. Of course, people immediately looked at what medical applications it could have.
There are dramatic medical examples where you're missing a key ingredient, or one of your ingredients is broken, when there is a disease associated with that - a mutation in a gene, or a missing gene, or duplicated gene or something like that. Cystic fibrosis is an example of that, where the problem is in a single gene. So there are definitely examples like that, conditions that can be identified, and understood in a certain sense by looking at this parts list, or looking at this ingredients list. But if you really want to know what's going on, in most cases a much more interesting thing to do is to look at the dynamics. That is in the proteins that are actually getting generated. Some of them are getting generated directly from genes, some of them are getting generated and then modified by after they are produced. There's a lot that happens after the genetics. And the proteins are controlling which genes are expressed.
So, to me, there is a much more interesting kind of analogy, based on process. The analogy we have so far is about structure. We emphasize the structure of things, so we think of the building blocks, and the things that get built, and the parts. I think it's much more interesting to look at the process that builds all of these parts.
It's true that the human body is an amazing structure, but what's much more interesting is the process that builds it, that maintains it, modifies it. That's not really in the genes, it's in the conversation that's happening between all the parts of the body, and the conversation is happening within the little molecular machines within the cell, or between the cells in the body. Your body has tens of trillions of cells in it, more than the population of the earth, and all these cells are talking to each other, sending each other signals, there's signaling going on within the cell.
To emphasize this other way of looking at it I like to look at the genome, not just as a parts list, but as the vocabulary list for this conversation. It's a useful thing to know, but the really interesting thing to do is to listen in on the conversation. What are these machines all saying to each other? That's what proteomics is about.
Proteomics is the study of all the proteins. "Omics" means "the study of all". The idea became popular when people like Wally Gilbert who started saying, "We should have all the human genes." Then by generalization, people were saying, "Well, we should know all the proteins in the body, we should know all the connections between neurons, and we should know all the metabolites." There are a lot of different kinds of "omics".
What is really interesting about proteomics, is the dynamic conversation; it's the study of the molecules that the genes are making, the ones that are controlling the genome. It is the conversation between the parts. This conversation is happening within the cell, and between cells, the elements of this conversation are proteins that are being sent around, and being absorbed by the cells, or being sent from one part of the cell to another. It's taking place in the medium of proteins, and so if you could see where all of those proteins are, and how they're dynamically changing, then you would, in fact, be listening in on the conversation. That would be a great thing to hear.
Biologists have recognized for a long time that it's a great thing to do. They've tried to do it. It's turned out to be technically much, much more difficult than genomics, for a couple of reasons. One of them is it's essentially an analog process, not a digital process. It matters how much of the protein is there. But another thing is there just wasn't this wonderful technology for dealing with it like replicating DNA.
You couldn't really do it well with the equipment that was lying around in the lab. People have tried to do it, but it was a very unrepeatable process, a very noisy process, and so the first publications about it tended to be wrong because people had mismeasured it, they couldn't measure the same thing a second time. So basically what happened was that it kind of got a bad name in biology, and people said, "Well, we can't get much useful information out of this", because, in fact, they couldn't get it with the stuff they had lying around the lab.
That's where I came in. I had looked at this in the abstract, years and years ago, I thought that this would be a great thing to do, but when I looked in to the details, I thought it would be too difficult. Then just a few years ago, I got approached by the oncologist, David Agus, who said, "We really need this information for treating cancer patients", and he convinced me to look at it again with the new tools that had come along.
The tools typically are things like mass spectrometers for weighing molecules, and liquid chromatography, which is basically sliding a molecule past a bunch of other molecules, and seeing how much it sticks. We can also make antibodies that stick to very specific molecules. That is a set of tools that hasn't changed very much, but when I started looking at it, I realized that the big problem was that people were using these tools basically in a lab bench, and treating it almost like they were treating genomics, as if it was a digital process. They were going through a sequence of experimental steps, but the way that they were controlling it wasn't possibly good enough to even get the same result twice if they measured the same sample, much less to look for subtle things in the changes.
I realized that it really needed a couple of other things, one of which is some better application of physics, which is how the instruments were actually tuned to do this problem. Another thing it needed was some plain process engineering. What was required was much more like making a semiconductor line than it was like sequencing DNA. There were many, many steps that had to be refined, and highly, highly controlled in order to get a repeatable result in protein. So this was essentially an engineering problem.
There are certainly hundreds of thousands of different protein variants, and maybe more. Nobody really knows which variations are significant. But certainly every gene produces a protein, and then those proteins get modified by the processes, and combined, and produce other proteins, and so on. The big problem was that there was no way of looking at all the proteins, say in a drop of blood that was repeatable, that you could measure the same drop of blood and see the same proteins, and part of the problem is because they occur in vastly different amounts. Some of them are a million times more diluted than others, so there's a huge dynamic range.
But also there are hundreds of steps in the process of measuring them. So if you're doing this with graduate students in a bio lab, and one of them goes and has a cup of coffee at one step, and you leave it 15 seconds longer than an enzyme, you get a completely different result. What needs to be done is a super tightly controlled engineering process.
Since that was essentially an engineering problem, I thought that could be an interesting problem for an engineer like me to work on, so I started working on that. Then it turns out that once you get that, there's a huge mathematical problem at the end, which I was also interested in. It was a computing problem, of interpreting all of these results. If you know that this protein is going up or down, how do you make any sense of that, and correlate that to anything useful. That is essentially a computing problem. Since it was a computing problem, and an engineering problem, I thought that I had something to bring to the table, and started working in the area really just to get the engineering worked out.
Applied Minds can do projects in the exploratory stage without going off and getting any funding; we do it with our own profits from other projects, so we started exploring proteomics with David Agus. And that's how we realized that we could do it if we could really build a line like an assembly line for doing it, which involved robotics, and changing the mass spectrometers, and things like that.
We got to the point where we actually knew how to do it, and at that point we raised some money from some angel funders, and made a company called Applied Proteomics, which has worked out how to do this, and built this assembly line which does these hundreds of steps, and measures along the way, and does it in an automated way. For the first time, the results are accurate and repeatable. When we test the same sample, we get the same result.
You can take a drop of blood, and get a repeatable measurement over a hundred thousand repeatable stable features. We don't know necessarily what all of them are, but many thousands of them we can identify as known proteins, and we now have genes associated with them. Often that means we know something about the function, or where they are created in the body, or something like that.
Let me show you the results of that process, which is on this slide.
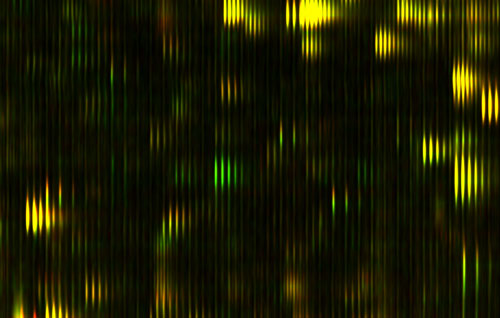
Figure 1: Differential Feature
This is actually a small part of the measure that we get out of a drop of blood; this is actually a small part of a bigger picture. We've spread out the fragments of proteins in two dimensions here. It's a little bit analogous to a gel that you might see, or a gene chip, the same protein feature will always appear in the same position every time. The brightness shows how much of that protein there is. This display actually doesn't show you too much of the dynamic range and brightness, but we are measuring that.
In the horizontal direction we're measuring the mass of the protein fragment. The vertical axis is how slippery it is. People have produced pictures like this before, but what's interesting here is that every time you do this, the features come out in exactly the same places. That hasn't been true before.
Just to show you how precise these pictures are, you notice that these things tend to occur in these little groups of stripes, tick, tick, tick. You see there are several of them in each group, and they kind of trail off; it's almost like a ring, or an echo. Well, the reason for that is that carbon has different isotopes, and so if there is an extra neutron, you have a different isotope of carbon in the protein, then it's going to be slightly heavier. That distance between the stripes is actually the weight of one neutron; it gives you an idea of how precisely we're measuring things. There's nothing in between because there's no such thing as half a neutron. In fact, measuring things so precisely we can often tell by the shape, how many carbon atoms there are in the protein.
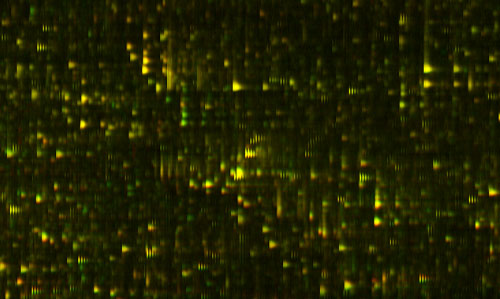
Figure 2: Yellow Overlay
The amazing thing about this picture is I'm actually showing you two measurements of two different blood samples on top of each other. One of them is shown in red, and one of them is in green. Things look yellow because the two are exactly on top each other, because the two blood samples are mostly the same. But, if I look closely at this ... actually let me just find a spot which is different ... okay, well, actually here is a good spot. There is some protein that was in one of the blood samples that wasn't in the other one, so you only see it as the green. There is another place where you see there's a little red down there. That is something that is in one sample and not the other.
It's almost as though we've got a digital read-out of this highly analog process. That's the amazing sort of engineering feat that I don't believe anyone else has achieved that kind of repeatable precision over that much range before. What we know is the relative concentration of each of those proteins. Now, this pair of tests might be the same person at two different times, or it might be two different people. They probably both have the gene to produce that protein, but for some reason one of them is saying this, and the other one isn't saying this at this time.
Now if we have a hundred thousand of these features — and we do have more than a hundred thousand — then the question goes on to what do they mean, what do we do with it? That's the stage that we're at now. It may be that some of those like a genetic test, maybe a single feature will actually tell us something. But probably much more of the information is in the patterns and combinations, and so on.
For instance, let's say that we go to cancer patients, and we try out a drug on them, and we find out that only 10 percent of them respond to the drug. It would be very nice, if there were some genetic marker that told us which 10 percent responded to the drug, because it's a miracle drug for those 10 percent, but it's a useless drug if only 10 percent of the people respond to it, and it makes 20 percent of the people sick. You would like to know which are which, and it was a great hope that maybe you would be able to find genetic markers to do that. There are a few drugs that that's true for, but by and large, that information doesn't seem to be just in the vocabulary list.
But the information is much more likely to be in here, there's something dynamically happening, and so if we can start to say, "If we see this pattern approaching expression, it means that you've got this thing going on metabolically." All of a sudden we've got hundreds of thousands of symptoms to look at, if you will, or hundreds of thousands of indicators of what is going on at the level of what's actually going on.
In the process, I started looking at how we treat cancer, and how we think about cancer. This is another area where I think there's a wrong paradigm that has gotten started because there's been a great success, and that success has been over generalized. In this case the great success has been the treatment infectious diseases and the germ theory of disease. This is the greatest success of a theory in medicine.
That was a very cool development, because if you can figure out what species of germ you were infected with, then that sets how you should treat the disease. You could treat the disease with something that would kill that germ. That became the general paradigm of medicine. You would do a diagnosis, a differential diagnosis to figure out what the infectious agent was, and then you would apply a treatment that was very specific for that agent.
That's the thing that doctors are basically trying to do, identify the disease, and treat the diagnosis according to the best method. That allows science to come in because you can objectively test whether a particular treatment is effective, or not effective, when dealing with that diagnosis. Does quinine help the symptoms of malaria? Is penicillin the best way to treat anthrax? Once you know what's best, that's the thing that doctors are taught to do.
Interestingly enough, that way of looking at things is not the only one in the history of medicine. Historically, doctors had theories that are today more like Ayurvedic medicine, with its emphasis on balances between various forces in the body. Or in the West, a medieval doctor might have tried to make you less choleric or more phlegmatic. The idea was to try to restore the order of the various forces that were controlling the body. It's interesting, at the time that the germ theory of disease was really exploding, and antibiotics were being discovered, J.B.S. Haldane said, "This is a disaster for medicine because we're going to get focused on these germs, and we're going to forget about the system." He was right.
Indeed, if you look at what happened, it was a disaster for treating diseases like cancer because we started thinking of them almost like they're infectious diseases. It's a habit of thought, so when a patient comes in, we diagnose them, and we put them in a category, and then we try to apply the treatment that is shown to work on that category. We do a blind clinical trial of how people that are in that category respond to a certain drug. That makes a lot of sense for infectious diseases because infections are species, they speciate, and divide out, so putting them in categories makes a huge amount of sense.
But a systems disease like cancer, or an auto-immune disease, is a break down in the system, much more like a program bug. We would never think of debugging a computer by putting it into one of twelve categories, and doing something based on the category. Actually we do, it is kind of "help-desk debugging" that doesn't work very well in complex situations.
There is a big difference between help-desk programming debugging, and the kind of debugging a programmer really does when they're trying to more subtly fix a program.
What we've got in medicine now is kind of help-desk debugging. We put you into a category. In cancer we start by putting it in a category that's based on the part of the body where symptoms of the cancer have been shown. Then we test drugs that way: Does this drug work on lung cancer, and if it does, well, it's not approved for prostate cancer because we tested it on lung cancer. That's a whole other experiment, that's a different category of disease. Then we subcategorize them. We take a biopsy sample, and we say, "Well, these cells are kind of squishy and long, and those are kind of round, so we have the squishy, long cancer, and the round cancer." We declare that we have two forms of breast cancer.
We keep coming up with more kinds of cancer as we measure more things, and then we subdivide the categories. There used to be dozens of kinds of cancer, and now there are hundreds of kinds of cancer. But I actually think there are millions or billions of kinds of cancer. Cancer is a failure of the system. Happy families are all alike, but unhappy families are all unhappy in their own special way, and happy bodies are kind of all alike, but when they break down, they all break down in their own special ways.
The breaking down is at the level of this conversation that's going on between the cells, that somehow the cells are deciding to divide when they shouldn't, not telling each other to die, or telling each other to make blood vessels when they shouldn't, or telling each other lies. Somehow all the regulation that is supposed to happen in this conversation is broken. Cancer is a symptom of that being broken, and so when we see a whole bunch of cells starting to divide uncontrollably in an area, we call that "cancer", and depending on the area, we'll call it "lung cancer", or "brain cancer". But that's not actually what's wrong, that's a symptom of what's wrong.
To use another kind of analogy, let's say we didn't understand anything about plumbing, but occasionally we came home and our living room is filling up with water, and sometimes we come home, the kitchen is filling up with water, and so we start describing the problem as, "Well, my house has water, that's the problem." We might even divide it and say, "My house has kitchen water, or my house has living room water." If plumbers were like doctors the best they might be able to say is "we've learned about kitchen water, and if we pour a lot of drano in the kitchen, then kitchen water sometimes goes away. Living room water is fixed by pouring a lot of tar on the roof." Indeed, there might be ways of fixing the problem, but what you really need is to understand about plumbing. You should be worried about the process that's creating the water, and understanding about what's supposed to be draining, and what's supposed to be holding it, and so on.
In fact, we misunderstand cancer by making it a noun. Instead of saying, "You know, my house has water", we say, "My plumbing is leaking." Instead of saying, "Somebody has cancer", we should say, "They're cancering." The truth of the matter is we're probably cancering all the time, and our body is checking it in various ways, so we're not cancering out of control. Probably every house has a few leaky faucets, but it doesn't matter much because there are processes that are mitigating that, by draining away the leaks. Cancer is probably something like that.
In order to understand what's actually going on, we have to look at the level of the things that are actually happening, and that level is proteomics. Now that we can actually measure that conversation between the parts, then we're going to start building up a model that's a cause-and-effect model: This signal causes this to happen, that causes that to happen. Maybe we will not understand to the level of the molecular mechanism but we can have a kind of cause-and-effect picture of the process. More like we do in sociology or economics."
Whatever the treatment of cancer, or auto-immune disease, neurodegenerative disease or other system diseases will be like in the future, there won't be a diagnosis step, or at least that's not what will determine your treatment. Instead, what we'll do is we'll go in, we'll measure you by imaging techniques, and taking it off of your blood, looking at the proteins, things like that, build a model of your state, have a model of how your state progresses, and we'll do it more like global climate modeling.
We'll build a model of you just like we build a climate model of the globe, and it will be a multi-scale, multi-level model. Just as a global climate model has models of the oceans, and the clouds, the CO2 emissions, and the uptake of plants and things like that, this model will have models of lots of complicated processes happening at lots of different scales, and the state variables of this model will be by and large the proteins that are moving back and forth, sending the signals between these things.
There will be other things, too. But most of the information is in the proteins. There will be a dynamic time model of how these things are signaling each other, and what's being up-regulated, and down-regulated, and so on. Then, we will actually simulate that under lots of different treatment scenarios; we'll simulate for your cancering, how we can tweak it back into a healthy state, having it guided back toward a healthy state. It will be a treatment that's very specific. We'll look at those and see which ones are most likely to bring you to a healthy state, and we'll start doing that, and we may treat you in a very different way than we've ever treated any other human before, but the model will say that for you that's the correct sequence to treat it.
Right now this would be a huge change in medicine. For instance, the way we pay for medicine is dependent on the diagnosis. You pay a certain amount for prostate cancer, and you pay a different amount for lung cancer. That determines what part of the hospital you get routed to, which doctor sees you, what the insurance company will pay for. If you take that out of the system right now, it's a completely different kind of a system. I don't think this will be an easy switch and I don't know what the sociological/economic processes will be. But it will happen because it will start working better.
It will probably happen first with desperate people who aren't getting fixed with the normal methods, will go to this alternate process, and when enough of them start getting fixed by this alternate process, then that will, by some complex sequence that I won't even try to predict, eventually change medicine.
Of course there is a lot to be done to make this work. We are dealing with very different time scales and different space scales, too. There are useful things that are hour-to-hour time scale, in your bloodstream, and your cell level is probably more like minute-to-minute, or even faster. Right now we're just beginning to be able to measure proteomics within a single cell, and so right now what we are doing with the National Cancer Institute is trying to bring all of those time scales, and space scales together into a model. This is probably, with today's technology, a ridiculous stretch, but we're at least attempting to do it.
We're measuring things both at the inside the cell level, measuring at gene expression the production of proteins, the placement of proteins within the cell, then the conversation between the cells. That we're trying to do at more the minutes time scale, and then we're doing what's happening in the body and the blood, on the days time scale. We should be probably measuring it over hours, but with current technology we can't afford to do that.
We're doing this in mice right now, but we can only draw so much blood from a mouse. Those kinds of things are limiting us. We're also doing it with imaging, the actual geometry of the tumors, so we're actually trying to measure geometry; we're trying to measure the genetic evolution of the tumor because the tumor is not homogeneous. Genetically it's different inside than outside, and we are trying to make a model, like one of those kind of global climate models, for lymphoma in mice. We can get genetically identical mice, and we can actually very reliably give them the same kind of lymphoma, and so we can repeat the experiments.
So it's much better than the global climate models situation. With global climate, we have one experiment to calibrate our model, and we're in the middle of it. There is no control. In the mice we can do a lot, we can try different variables, and then also try what are the effect of different things that we can do in terms of treatments, of giving chemotherapy of various sorts, or heating them up, changing the pH of their blood, doing all kinds of things like that, and begin to get a perturbation model of not just how the system normally works, but how it works under different kinds of perturbations. Then hopefully, eventually, we'll get to the point where we have a good enough model that we can actually predict: if we do this to this mouse we can actually make it live longer.
We are already learning a lot. One is, for this mouse study, we're combining some new techniques like proteomic techniques with a lot of techniques that were developed for other reasons, for instance imaging techniques that are very detailed. We're actually putting little windows into the mouse, and watching the tumor grow, and then we can use things that have antibodies that bind to certain kinds of proteins that are being expressed, and so we can actually see where those proteins are being expressed in the living mouse, so see geometrically where they're being expressed.
There are techniques, for instance, where we can actually look within a cell, and see where a protein is within the cell. We can actually do microscopy below the wavelength of light now, which is a fantastic advance, by using basically little flashes of light, and computing on top of it. There are huge advances in the technique and instrumentation and so on that's making this at least conceivable for the first time. It's only a matter of time before it will be possible, and it's quite probable that this first attempt is too early, but we are attempting to do this with the consortium of people in places like Stanford, and Cold Spring Harbor, and USC, UT, NYU and Caltech. The National Cancer Institute, has actually given our group five years of funding, assuming we keep making progress.
They had this crazy idea of getting people like me, who are not really biologists, to be the principal investigators of these centers, to work with clinicians to design the program of research, which is then being carried out by a lot of people who know things like how to put windows into mice, and how to image a tumor, or how to get antibodies to glow. So we're using all of those biological lab techniques to do something that's really more like a physical sciences model.
I'm optimistic that we'll have enough success that people will at least try to repeat this form of experiment. Whether we actually are able to make accurate predictions is yet to be seen. The group coup would be if we got to the point where we could say, "We can predict that if we do "this" to this mouse, then we can take care of "that" in the mouse". That would be success. But we can learn a lot without getting that far.
![]()
CANCERING: Listening In On The Body's Proteomic Conversation (PART II)
W. Daniel Hillis
What I've been talking about here is more analysis than construction. The genome is used to construct things, and I'm claiming it's not the best place for analysis of what's going on. Certainly there are times it is useful, but I don't think that's where most of the information is. In fact, in some sense, it is literally true that the information that's in proteomics tells you everything that was in the genome, everything useful that was in the genome. In a sense, the genome is redundant if you have the proteomics, that's theoretical though, because the genome is digital, and we actually have it. In many ways it's enabled proteomics. ...
Text (Part II) [12]
Printable text file (Parts I & II) [11]
(During this session Hillis was asked to comment on a number of specific topics:) CANCERING: Listening In On The Body's Proteomic Conversation (PART II)
W. Daniel Hillis
On the relationship between genomics and proteomic testing
What I've been talking about here is more analysis than construction. The genome is used to construct things, and I'm claiming it's not the best place for analysis of what's going on. Certainly there are times it is useful, but I don't think that's where most of the information is. In fact, it is literally true that the information that's in proteomics tells you everything that was in the genome, everything that was useful. In a sense, the genome is redundant if you have the proteomics. That's theoretical though, because the genome is digital, and we actually have it. In many ways it's enabled proteomics.
Right now, when I show you that image that has the hundreds of thousands of dots on it, I can actually tell you what a lot of those things are because we know the genome; so I can actually associate many of those dots with genes, and because we have these great genetic expression tools and so on, we may know what part of the body it's in, we may know a lot about the pathways because we can do knock-out genes. It's a great experimental method for actually controlling what proteins get produced and so on. In many ways proteomics was made possible by genomics. It builds on top of genomics. I guess it's true in some theoretical sense that eventually you might not even bother to look at the genome if you can see the whole proteome. In practice, it's been very important.
The genome is the instructions for the cell. That's very important if you want to do manipulation. If you want to actually affect the pathway, then that is the level at which you need to manipulate things. You want to knock out a gene, or modify a gene. Experimentally, being able to read and write the genome is incredibly important. But if you want to use it as a diagnostic for what's going wrong with a particular individual, it will be unusual for that information to be in the genome.
On the role of gene testing in cancer
Let's take somebody who has cancer. They used to be somebody who didn't have cancer, and they had the same genome. So the difference between having cancer and not having cancer is clearly not just in the genome. There's more to it than that. In fact, most of their cells aren't cancering, and they have the same genome. Cancer is a dynamic process that's happening, and it's not just in the genome. Now, there may be a specific mutation from a genome that helps explain why it happened. For instance, one of the dramatic genetic test successes has been in breast cancer, BRCA1 and 2, which are specific genes that are associated with breast cancer. They occur a lot in Ashkenazi Jews, and a particular kind of breast cancer is associated with these genes. There are many examples like this where there's a genetic predisposition to cancer, but is one of the clearest examples. The cancer isn't inherited, but that's a predisposition that is, so people that have the gene are more likely to have cancer.
Let me put it in terms of the conversational analogy. That means that certain words are missing, and in that case we know there is a conversation about fixing broken DNA. We're repairing broken DNA and it's hard to describe what to do without those words. You need to discuss BRCA1 and BRCA2, and you need to use those concepts. You need to use those words in order to repair DNA in a certain way. If you don't have those words in your vocabulary, then you're unable to execute this process of repairing DNA.
If those words are slightly mutated so that they're not understandable, if you slur them, or stutter them or something like that, they're won't have the desired effect. It won't always cause a problem because there may be other pathways that also repair the same defect. If the other pathways are working very well to control your DNA, it might not matter. But there is an association, and you can look at people, you can test them, and you can say, "Well, if they have this mutation, they're more likely to get breast cancer", and actually, it turns out, they are much more likely to get breast cancer.
Furthermore, you know what the failure mechanism is, so there are many people who get breast cancer for completely different reasons that have perfectly intact BRCA1 and BRCA2. By the way, that same pathway, because it's a general pathway for repairing DNA, is also important for ovarian cancer because if you don't repair properly, then you're more likely to get ovarian cancer, too. It doesn't really have anything much to do with breasts other than that's where you see the symptoms of this happening, it's usually first noticed in breasts.
On the application of proteomic modeling to cancer treatment
The cancering metaphor does mess up our standard model of medicine, where we just take the right pill to fix a given problem. But if you think about it, the idea that you should be able to take a pill, and it should magically fix a disease, a systems disease, a failure of the system, is kind of amazing that that's even possible. The cases where it's mostly possible is where you have an invading thing that doesn't belong, like an infectious disease, and you take a pill that poisons that particular thing, like an antibiotic. There are a few cases where you're just missing one component, and you take a pill that provides the missing ingredient, and so there will be a few magic cases like that, but those are very special kinds of failure, and I don't think they'll be the typical failure in cancer.
Unfortunately this whole idea of fixing a disease with a pill, while it's delightful when it works, is not very generalizable. We haven't found very many new pills lately that really cure diseases. In fact, the pharmaceutical industry is kind of broken right now because they've run out of this low-hanging fruit, a magical chemical that cures a disease. I don't think we're likely to find a lot of more those. They need a different model.
The first commercial application of proteomics will be diagnostics and probably something much simpler than what I was describing before. The way proteomics will get started is by being markers for diseases that we already diagnosed, in other words, it will help support this categorization system of diagnosis.
Let's say I could find a pattern of proteins expressed in your blood that said whether your colon was growing polyps in it. That would be much better than having a colonoscopy. You get a colonoscopy every five or ten years, which is now recommended — even though in one in a thousand people they create serious damage giving the colonoscopy — we still recommend that people do it because we don't have a better way of telling if you're in this kind of precancerous stage. If you could do that in your annual blood test, it would be much better. That is an example of a very early use of proteomics. Probably there would be many things like that where if you could detect breast cancer without a mammogram, or if you could do a confirmatory test of prostate cancer without doing a needle biopsy, all of these things are very invasive, and expensive, and cause a lot of secondary harm to people.
On the relationship between proteomics and synthetic genomics
At last year's Edge Master Class in Los Angeles, George Church and Craig Venter talked about Synthetic Genomics. Proteomics is relevant to that because it's also a tool that such researchers could use since they need to debug their synthetic genomics. When you write a computer program, the first thing you do is you try to run it, and it almost always has a bug in it, so you see what happens, and you debug it, you stop it in the middle of running, and you see what the state of the system is, and you understand what your bug is, and then you change the program.
Right now George and Craig don't have the debugger. Proteomics is the debugger that they need. When they write a program and it doesn't work, which actually happens a lot, in order to tell why it's not working, they need proteomics to say, "Oh, I see, this isn't upregulating that enough, or downregulating that ... ", and that will help them debug their program and tune their program. Right now a lot of synthetic genomics is about copying a naturally-evolved program, and saying, "Okay, I can make a copy of this program, and write it, and it does the same thing", and that's interesting. It would have been very surprising if it hadn't worked.
On the business of proteomics
The business of proteomics had a false start a few years ago. As I said, proteomics can't be done with the techniques and tools that are sitting around a laboratory, a biological laboratory; it's not a biology problem. Unfortunately there are a lot of people who tried to do it with those tools and so there were a few companies that started up, and a lot of laboratory projects that started up, and they published a bunch of results probably prematurely, and they couldn't replicate them because, you know, the next time they ran the tests it came out differently. It was so noisy that they had to do very, very large trials. So if you have a bad instrument, then it's not going to work.
Because of that, proteomics got a bad name among the venture capitalists. Probably most venture capitalists will cut and run if you say "proteomics" right now because of the problems with the tools of a few years ago. What will happen is we'll get some successes. In spite of this, there will be a few things like Applied Proteomics that get started, and there will be a few people with more vision that look at it more closely, and say, "This actually fixes the problem that they had before, because the story was right before, it was just they couldn't make it work." As soon as there is a success, then you'll see a general change in attitude. Venture capital tends to work this way. There will be lots of people investing in this area, and there will be a boom very much like there was in genomics, and sequencing. Lots of effort will go into both the technology of doing the proteomics better, as it happened with genomics, and also the application of proteomics.
And again, first it will be to these diagnostics, and then it will be things like drug rescue. Billion-dollar drugs that had to be taken off the market after having had huge amounts of money invested in them because one in ten thousand people reacted badly to them. Well, if you could tell who was going to have that reaction, if you could make a test for it, and again, if you could find in a proteomic marker for what it was about, what was wrong with the dynamics of their body to cause them to have that reaction, then all of a sudden such drugs would be viable again. They could recover that billion-dollar investment. You'll see pharmaceutical companies take it up for reasons like that. That's the "safe" part pharma companies are looking for drugs that are "safe and effective", so it will help them with safe.
It will also help them with "effective" because right now, if you have something that not everybody responds to then it is not effective. We've had the first examples of that already, where a drug wasn't statistically effective. But when we look again we can say, "Well, people who have this protein being expressed, it is effective." So there is a very simple case where you're looking at a single protein. This single-protein expression is a situation analogous to a conversation with somebody shouting "Fire". A single protein is a very degenerate case of the conversation. There will be a few markers like that, that are simple markers, but mostly I think we'll look at things that are much more complex patterns that happen in multiple proteins.
Dosage is another reason why pharma companies will get very interested in proteomics, and it probably will make a big difference in research. Right now you can't tell what's happening until it gets all the way up to having a symptom in a patient. If you have something that takes a long time to play out, like Alzheimer's disease, or ALS, you really don't know if your drug is doing any good for years and years. You have no idea if the dosage is too high, or the dosage is correct. The feedback loop is very, very indirect, and lots of other things are affecting it, too. But if you could actually look at the proteins and notice this bad problem of communication between the cells, that's causing plaque formation in the brain in the case of Alzheimer's, then you might be able to see the response of the drug immediately, even though the symptoms aren't changing yet. The symptoms may take years to change, but you can see the drug is effective in this patient immediately… or that it is not, so you can move on to trying the next drug.
You could titrate the dose and very quickly say. You can titrate the dose not only so the guessing holds the correct, safe dose, but actually specifically for the patient. We know that people respond very differently to drugs. Right now it's a trial and error process. They give you a little bit, if that doesn't work they give you more. We see this happening even with simple drugs like blood pressure drugs. Now, with blood pressure, doctors can measure blood pressure easily. They should give you a small dose of the drug first, they see if your blood pressure went down enough, and they can give you a little bit bigger dose. You can do that as a quick loop and you calibrate the response for an individual because you can measure blood pressure. Well, you can't do that for something where the outcome is years away. Proteomics ought to let us do that, too.
On the way proteomics will change treatment
Something that I skipped over in describing the new treatment paradigm, and the simulation paradigm of treating rather than the diagnosis paradigm, is that right now what you do is you diagnose, and you select a course of treatment. Now, what really happens if it doesn't work, maybe you switch and jump on to a different track. But the interesting thing, once you can kind of measure the dynamic state variables, is you're constantly redoing that. You replan every time you go into the doctor. There is not a notion of staying the course, there's no reason to stay the course because you can tell you're getting closer. You redesign a new treatment every time you remeasure. You're constantly getting feedback. You don't just design a special customized treatment per person, you do it per person per time, per person per visit to the doctor.
What you're trying to do is guide it back to a healthy state, basically. But the other great thing is because you're looking at the whole state, you're not just treating one thing at once. That's the other thing that's a flaw in this idea we've gotten about medicine, which is in the infectious disease model. The fact that you have malaria is the main event. You've got to kill off that malaria before anything else can happen. But in the systems diseases, there's lots going on in the body, constantly. So when we say a treatment is safe and effective, the way that we test that is we're testing one thing at once, not looking at everything else. So we say, "Okay, well, some statins lower your cholesterol, which we think is a surrogate for you're going to have a heart attack", so we give everybody a statin.
Now, if you look at some of the early monkey studies where they first studied statins, and proved that it lowered cholesterol, the monkeys that were taking the statin had a higher death rate than the monkeys that weren't taking it. What does that mean? Well, they kind of ignored it, because they looked at the deaths and they were from accidents, or getting into fights, things like that. I bet that it's going to turn out that statins actually cause you to get into fights, and have accidents. It will have effects on your mind that weren't studied in all the studies of statins, and in fact, statins may not be good for you. Or they may be good for you for reasons that have nothing to do with lowering cholesterol.
That's another thing that we're discovering in statins. The idea that we go in there, and we have this very complicated system, and pick one variable and say, "Does this one variable get better when we give this person this pill?", and then say, "Okay, well, it did." The engine rate went up because we poured this goop into the engine. But is that really good? Well, maybe the speed went up because it broke the regulator or because it clogged up the safety valve. So right now when we test the drug, we're looking at one thing at once. We only discover these bad side effects in these retrospective studies looking back after people have been taking the drug for a long time. ?
What most of these drugs do is shift the balance. They're trade-offs. There is a reason why you don't naturally produce statins. I don't think it's because nature didn't think of the idea, it's probably because statins have some pluses and minuses, and so what you're doing is you're rebalancing things, tipping things in a certain direction. The reason that's relevant is if you're in the paradigm of diagnose the disease, treat the disease, then that tends toward treating the disease at the cost of making everything else a little worse, than if you're looking at the whole system, and trying to optimize the whole system. Proteomics is looking much more at the whole system.
Proteomics can also tell you things that can give some substance to what nutritionists have been talking about. If you look at traditional Chinese medicine, or Ayurvedic medicine and things like that, it's all in terms of balances, restoring this force against that force, but it doesn't have a very good model. It is a highly oversimplified model that's been made up without much scientific information going on, just a lot of time going into it. If you had a much better model, then you could probably rationally understand what foods you should really eat that bring your body back into balance.
It is probably very different for different people, and so I don't think the treatment is necessarily going to be taking a pill. You will take pills, but you'll also change diets, or you may discover that it's very important for you not to be stressed out in the afternoon, you may find it's important for you to get lots of oxygen, or do something very aerobic. You may find out that you better not get cold. Or we may deliberately heat you up to an abnormal amount for a little while. Maybe you will need sequences of these things. I don't think treatments will be "a thing", like a pill, I think treatments will be perturbations, bringing you toward health, with some feedback as to what results all those perturbations are having.
In a sense we already have home proteomic tests. Some of these home pregnancy tests may be that. What's happened is, for various reasons, we have luckily discovered certain proteins that are diagnostic, PSA is a great example.
PSA is a protein, a prostate-specific antigen, and for various reasons we happened to notice that this was a marker for prostate cancer, that it was associated with prostate cancer. Now when you go get a blood test, if you're male you probably automatically get a PSA test. That is a protein test — a test for a specific protein. Once you can look at the whole protein, one of the things that will happen early is you'll identify specific proteins to test for, and those may be home tests. They may be finger prick tests, or there may even be indicators of a protein that are excreted in your urine, so it wouldn't surprise me at all if you get home tests that come out that were identified by looking at the whole proteome. But my guess is, again, those are going to be kind of unusual, special cases.
In some sense it's really lucky that there happens to be a single protein that's associated with inflammation in the prostate. In fact, maybe it's lucky, maybe it's not, because there are a lot of people getting needles stuck in them unnecessarily because of this test, and probably they would be much happier not being told they have prostate cancer, because it probably won't hurt them anyway. Maybe the needles are worse than what would have happened without the test. It causes a lot of worry, and stress, and maybe the stress even makes them prone to some other kind of cancer.
Probably there is a much more sophisticated combination of lots of different proteins that you can look at and say, "This is an inflammation of the prostate that's actually not cancer." Or, "This is an inflammation that looks like this particular kind of cancer that responds to this drug, while this is a particular kind of cancer that grows very slowly, and it's not going to kill you. You'll die of something else first, so you don't need to worry about it." Once we get much more subtle measures, once we can listen to that conversation and find out what's actually going on, then I think there will be a lot less treatment. We do a lot of overtreating of things, and a lot of damage by treatments. That's actually one of the only things that can actually reduce the cost of medical care. The cost of medical care is mostly way down the line when people at hospitals, are already pretty damaged by disease and treatment by the time they become very expensive.
If what you can do is spend more of the money up front diagnosing things, tweaking things before it gets to that stage, then that is a win-win because it's better for the patient to get less treatment, and better for the insurance company because you never get to the hospital. Of course, the doctors actually are happy because they really do want to cure the patient. They have more tools to actually know what's going on. It's very frustrating for the doctors to know that a treatment might not work.
Doctors hate this thing of giving you a sequence of poisons: hopefully we'll kill the cancer faster than it will kill you. That's a horrible thing to do to a patient. I think oncologists would love to have better ideas of what the effects of the treatments that they're giving are, and is it actually doing any good. They want to avoid giving people treatments that are not going to help, that are going to make them sick.
On home testing
I'm sure we'll have examples where people can do self-testing, perhaps for a single protein. I think it's going to be rare cases. Sometime we may see proteins or parts of them in the urine. Mostly proteins get broken down, unless you have something like bladder cancer.
Usually we test the blood. But, diabetics do blood draws all the time at home, and tests. So I can certainly imagine things that are the equivalent of diabetic sugar meters, glucose meters that you're adjusting your own dose of blood pressure medication, or anti-cancer medication, or something like that. Right now proteomics makes it very expensive to do because we're just able to do it for the first time. But certainly once you've identified what you're looking for, then there is some fairly standard methods of changing that, and turning that into tests that are very easy. The classical method is you develop an antibody to the protein, and then you can make something that changes color. More and more it will be changes to the conductivity of a transistor so that you won't read out a color, you'll have a little machine that does the analysis, not just indicating presence or absence.
I think the interesting information will be in the levels of many different proteins. The picture I showed you was a protein that was produced in one and not in the other. Much more common, though, we see proteins that change in levels by 20 percent or something like that. A lot of the information isn't just on/off, it's in the concentration. You don't totally turn it on or off, you change the rate at which you're producing it and a lot of drugs basically do that, or they change the rate of production or destruction of something. They upregulate something, downregulate something else. If you take BRCA as an example, there's another protein that I think regulates a promoter of BRCA production, and actually if you have a mutation in that, too, then the mutation in BRCA is less important because it's not regulated. That's a completely different kind of subcategory of breast cancer.
When you try to downregulate something, it is because you have too much of it, so you lower it by giving a drug that suppresses its production or speeds up its destruction It's all in the quantities, it's not just present or absent like it is in the genome, and that's the thing that's really neat about the genome is it really is digital. You either have a gene, or you don't. You have this variant of the gene or you don't. You have one or two copies of it.
I'm emphasizing blood because it's nice you have a collecting system built into your body that goes around and touches every place in your body and collects fluids. It's very convenient for diagnostics. But there will probably be things that happen in tears, or in saliva, or in lymphatic fluid, or spinal fluid, but your body is plumbed very nicely for blood so that you can get it out easily, and since it delivers nutrients, and gets rid of waste, it pretty much is involved in anything very dramatic that's happening in the body. It's good low-hanging fruit, at least.
In diabetes treatment, what we do now is we basically get around your pancreas. Doing what your pancreas does by adding insulin. We try to mimic the pancreas. Proteomics may allow us to actually stop the processes that cause your pancreas to go bad earlier. We know that that's something that can be affected by diet. Again, it's probably predispositions, so if you're missing certain vocabulary words, it's harder for certain kinds of processes to work, and so you are particularly susceptible to certain kinds of problems, so that's where genetics comes out. Then proteomics can see, are you actually having those problems that you're susceptible to, or are your other redundant mechanisms taking up the slack for it? The body seems to be highly redundant, and often has lots of mechanisms for doing anything important.
On redundancy and treatment
Probably the biggest surprise that happened with genetics is once we had the ability to knock out genes, everyone thought, "This will be great, we'll knock out the gene and see what breaks." Well, it turns out mostly when you knock out a gene, at least half the time nothing seems to break. There was some redundant thing that just took up the slack. Probably if you did very careful studies on those knock-out mice you would find out that they were more susceptible to certain kinds of diseases or breakdowns because they're missing one of their redundant mechanisms.
It's interesting that evolved systems, much more than engineered systems, are evolved to be robust and redundant, and if you think about it, robustness is a kind of information hiding. What you wanted to do is you wanted to respond the same under many different kinds of circumstances, and with some of its parts broken. What you're doing is you're hiding the information in effect about, how it's working. The very process of making something robust is a process of hiding the information at the level of the symptoms, at the level the physician is looking at. In a very real sense, evolution has evolved your body to make the systems uninformative about what's going on inside.
It's kind of amazing that a doctor can look at things on the outside of you, and take your temperature, that it works at all that you can look at the outside of somebody and decide what you should do to them to make them better because evolution has evolved it for that not to work, in effect. It's actively hiding the symptoms of what it's doing, because that's what it's evolved to do, it's evolved to do the same thing no matter whether it's working or not in some sense, or whether each piece of it is working or not.
You do need to get to a different level to see what's going wrong. Mostly the body fixes itself when it's cancering, and so if you can just sort of help it, tilt it in the right direction, probably most cancers would fix themselves if you give them a chance to.
Oddly, though, if you think about that and think about chemotherapy, which is basically the strategy of let's try to poison the cancer, even though we poison you, it seem crazy. Let's try to have a poison that's just enough not to kill you, but to kill the cancer. Well, you're also probably compromising all those mechanisms that fight the cancer, rather than propping them up. In some sense we're probably making the body's job harder in our attempts to treat it.
Mainstream medicine tries to understand what are the mechanisms, what are the pathways, tries to design things that enhance, or block something bad from happening and so on. Proteomics is a new tool that gives you a whole lot more information to do that with, and once you have a whole lot more information, then that lets you do a much more informed dynamic intervention. It helps to give you insight in the actual works of the pathways. That's a very mainstream idea. I don't think anybody would argue with that.
On symbiosis
A really interesting point is that probably a lot of the system that is our healthy body actually doesn't have human DNA at all. It has some other microbial DNA, and we probably are a complicated ecosystem of different types of our own cells, and lots of non-human microbial cells. Once we start looking at the proteome, we'll be looking actually at the conversation of all of those cells, not just the human cells.
Some of those stripes that show up in the picture I showed, we can say, "Oh, that corresponds to this human gene." Some of them, we don't know they might be produced by some combination of other proteins, might be some other organism's protein. Microbial protein would show up in that too, or response to microbes. One of the beauties of this is that we see everything, whether it's of human origin or not. We actually can see it all in the proteome.
On the National Cancer Institute
The leaders of the National Cancer Institute are very keenly aware of how little progress has actually been made in the treatment of cancer. This is something they pay a lot of attention to. They're thinking very laterally in giving funding to people like me to work on cancer. What they've done is to say 'Let's bring in some new kind of thinking to this, and let's have a program where we have physical scientists be the principle investigators, partnered with the co-investigators who are clinicians and biological scientists. I'm partnering with David Agus, for example. But giving money to the physical scientist is a pretty radical idea; you can imagine it is very controversial within the biological community. NCI has started start a few of these centers, and given them five years to work. They need to be interdisciplinary and geographically distributed.
Our center at USC has people all over the United States involved in it, like Cold Spring Harbor, Stanford, Arizona, UT, NYU and Caltech. Typically these labs will have a particular technique for studying biology. It might be measuring proteins within a single cell, or imaging the growth of a tumor. Very often these things are very, very hard to do, and so people have built their career on a new way of sensing proteins inside the cells, and they have teams of graduate students who are refining this technique, and doing variations of this technique, and developing it and so on.
There is about a dozen of these physical sciences centers, each of them is the same kind of structure, there is a physical scientist who is paired with an oncologist, and a group of people that are geographically distributed, applying their techniques to a program of research. In our particular program of research, as I've described, we're trying to take one cancer, study it with all these different techniques, and build a model of how it develops. Different centers have different goals for what they do, but what they all have in common is somebody who is not a biologist trying to use the tools of biology to study cancer in some way.
The proposals for these centers got reviewed by a peer review process, with biologists evaluating at them. One of the reasons that the biologists liked our program was that, surprisingly enough, it's really unusual, even unique for somebody to take one line of cancer and study the same line of cancer with all of these different tools. Typically what people do is study a different cancer with a different tool, and they find the cancer, the type of cells that they can grow in that lab that works well with their tool. The idea of picking a few different specific cell lines, and studying them from a lot of different dimensions, and lots of different scales; I believe that, our proposal probably got funded because the biologists said, "That's going to be useful even though we don't believe this modeling stuff will ever work. Just doing that will be very useful."
A lot of the resources that have been put into cancer have generated the preconditions so that we can do this. I don't feel those resources were wasted in any way. We really understand a huge amount about pathways, and specific mechanisms, so we know a whole lot more about cancer, even though we can't treat it much better in many cases. We have a lot more information, and that information can be made more useful now that we have this proteomic information. This isn't in the category of throw away everything we've done and start over, this is an incremental progress next step that's built on top of an incredible mountain of work.
On how these efforts could fail
There are a couple of ways that I could be wrong, or fail. The timing could be wrong. It could be that we don't have enough of that information built up about the mechanism to interpret these results yet, so it could be that when we really started analyzing we just failed to find the patterns of proteins that correlate with useful things to treat. I would bet then that that's probably just an issue of timing. We're too early, we don't have the right information to bring together with it, or maybe we're not doing the measurement in enough detail.
Maybe it's not these 20 percent changes, maybe it's 2 percent changes, or maybe it's changes in much rarer proteins that we're not measuring by this method, something like that. Maybe it's not in the blood. Maybe most of the information stayed inside the cell. Maybe the action is happening within the cell, not between cells, in which case, probably it will be much longer before it's diagnostically useful in medicine.
Eventually, almost certainly, it will be right. Whether, the timing is right to actually apply this information to medicine in the near future is a risky proposition, and the answer is probably they'll be partially right. There will be some things you can apply it to, and some things that remain mysteries until you look at much more detail of what's going on within each cell, or something like that.
Certainly there are other kinds of molecules that are important, too. It's not just proteins. We know, for example, glucose is important. There are small molecules that are important. Now, most of the small molecules, their production, breakdown, are regulated by proteins, so I'm guessing that if you have all the proteins' state, then you can infer what's going on with the other stuff. But there is state in the body in other things besides proteins. But most of the information seems to be in the proteins.
The body has lots of mechanisms for not only transmitting these proteins, but listening to them. If you look at what these molecular machines do, they talk to each other through these proteins; they say, "If you turn on, produce more of that, or less of that". They're measuring these proteins all the time, and responding to them. That's how your body works, that's how the feedback loops work. Your body is doing proteomics constantly.
It's exciting because for the first time we're really looking at the variables of this complex process, the dynamic variables, which is what life is. That may be too complex for us to comprehend, but at least we're listening in on the conversation. Whether we can understand it or not, we'll start learning in the next few years.
